5
New Biological Measurement Opportunities
INTRODUCTION
Recent and forecasted advances in microbiology, molecular biology, and analytical chemistry make it timely to reassess the long-standing paradigm of relying primarily or exclusively on traditional microbial (predominantly bacterial) indicators for waterborne pathogens in order to make public health decisions regarding the microbiological quality of water. This chapter provides an overview and discusses various issues and methods for making biological measurements. It underscores some of the key issues in making measurements both generically and specifically for pathogens and indicators of waterborne pathogens. The methods are evaluated critically in terms of their attributes, including potential applicability for measuring indicators and pathogens, as well as their limitations. The issues of standardization and validation of methods are then discussed, followed by a look toward the future that describes how new and emerging technologies and science will facilitate waterborne pathogen and indicator measurements. The chapter closes with a summary of its conclusions and recommendations.
Spatial and Temporal Granularity
As discussed in Chapter 4 and illustrated in Figure 5-1, the spatial and temporal scales (i.e., the “granularity”) at which indicators and indicator organisms are employed may differ widely among applications. Small spatial and short temporal scales (area A) are of particular interest in beach monitoring programs and,
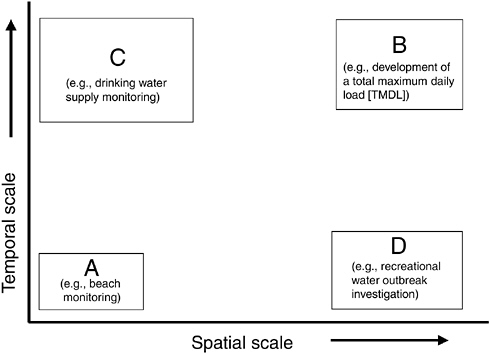
FIGURE 5-1 Spatial and temporal scales of indicators for various applications.
potentially, to transient contamination of groundwater. Larger spatial scales and longer temporal scales (area B) are of importance in understanding overall sources of microbial loadings to a watershed (that may serve as a water supply) or in studying the contamination of an aquifer or well. Small spatial scales but long temporal scales (area C) may be useful in understanding “typical” conditions at a water supply intake on a river system for the purposes of developing treatment configurations to meet drinking water standards for finished water. Large spatial scales (area D) but short temporal scales may be useful in understanding the occurrence of contamination over a large recreational area under outbreak conditions or from a storm event.
The temporal and spatial requirements for each particular application largely dictate the types of indicators or indicator approaches employed and the methods for measuring these indicators. As discussed throughout this report, particularly in Chapter 6, what is needed is a phased monitoring approach that makes use of a flexible “tool box” in which a variety of indicator methods and approaches are available for measuring a given indicator or pathogen for differing applications and circumstances. In many indicator applications, the level of perceived public health threat will determine the method or methods employed, as well as the spatiotemporal granularity. The indicator method, frequency, and spatial coverage of sampling will have to be “adaptive” in the sense that more frequent samples
taken over larger areas with more sensitive methods will be required when the threat level is high (e.g., following high rainfall events) than when the threat is low. In some cases, the number and type of indicators measured may also differ with changing environmental conditions.
Classical Methods and Their Limitations
Most of the indicator applications described in previous chapters rely on biological measurements of bacteria. The classical laboratory techniques presently used for those measurements are primarily culture based, involving quantification of a metabolic or growth response after a suitable incubation period in an appropriate substrate. As reviewed in Chapter 1, culture based methods have been used for more than 100 years in water and related areas of environmental microbiology and have been considered adequate to provide quantification of indicator organism (predominantly bacteria) concentrations. Culture methods may be limited by their incubation period since most require 24 hours or longer, during which time the public is potentially exposed to a health risk (see Chapter 4 for further information).
The current choices of detection methods for indicator bacterial species or groups were motivated by the associated technical difficulties in culturing many types of waterborne pathogens. However, it is now possible to detect the growth of some specific pathogenic as well as indicator bacteria and also some viruses and parasites in as little as a few hours. For example, in clinical diagnostic and food microbiology bacteriology, automated bacteria culture detection and identification can be achieved in four to six hours (Fung, 2002; Lammerding et al., 2001; Murray et al., 1999); however, these and other advanced methods for rapid culture detection have not been well developed for or adapted to the rapid detection of indicator or pathogenic bacteria in water and other environmental samples. One reason why rapid culture-based detection works well in clinical diagnostic microbiology is that clinical specimens often contain high concentrations of the bacteria of interest, thereby allowing them to be cultured to even higher concentrations in only a few generations. In contrast, water and other environmental samples often contain very few bacteria of interest and therefore, many generations of bacterial growth are needed before these bacteria are readily detected by culture methods. Besides bacteria, coliphages—which are bacterial viruses infecting Escherichia coli (E. coli) that have been shown to be useful microbial indicators of fecal contamination and predictors of human health effects from recreational water exposures (see also Chapters 3 and 4)—can be cultured and detected in as little as six to eight hours by some methods (Lee et al., 1997; Sobsey et al., 1990).
As discussed in Chapter 3, many types of pathogenic and indicator bacteria present in the environment are in various states of physiological health and fitness, depending on their origin, properties, and how long they have been in the
environment. The state of the microbes is influenced by the extent to which they have been exposed to various environmental stresses such as extreme temperatures and pH levels, hypo- or hypertonic salts, aerobic or anaerobic conditions, UV radiation, heavy metals, and various other antimicrobial chemicals, including chemical disinfectants such as chlorine (Hurst, 1977; McFeters and Camper, 1983; McFeters et al., 1986a,b). Therefore, enteric bacteria and many other bacteria in aquatic environments that are stressed, injured, and physiologically altered, may or may not be detected by various culture methods (Edwards, 2000).
Typical culture methods for pathogen and indicator bacteria in water and other environmental samples greatly underestimate the true concentrations of viable and potentially infectious cells—sometimes by as much as a thousandfold (Colwell and Grimes, 2000; Ray, 1989). For example, the anaerobic enteric bacteria that are so plentiful in the human and animal gastrointestinal tract, such as Bifidobacteria and Bacteroides (see also Chapter 4), are very difficult to culture from water and other environmental media because they are highly sensitive to very low concentrations of oxygen. While these bacteria would appear to be attractive candidate indicators of fecal contamination, the inability to culture them efficiently from water and other environmental media has been a major impediment to their potential use as fecal indicator microbes. However, the advent of nucleic acid based molecular methods to detect these bacteria now makes it more plausible and practical to consider them as fecal indicators (Barnhard and Field, 2000).
The underestimation of bacteria concentrations also results in part because the differential and selective media used to culture many types of waterborne pathogens and indicators contain inhibitory agents intended to suppress the growth of nontarget bacteria. Such agents also suppress the growth of injured or stressed target bacteria. In addition, other culture conditions, such as elevated incubation temperatures, may contribute to the lack of growth of target bacteria. Because bacteria injury is induced by the chemical disinfection and other treatment processes applied to water and wastewater, McFeters and colleagues (1986a,b) greatly improved the detection of injured coliform bacteria in water (by more than 10-fold) by the use of a medium that contained fewer inhibitory ingredients. According to some authorities, such bacteria can become viable but nonculturable (VBNC), as discussed in Chapter 3 and below.
Whether the VBNC pathogenic and indicator bacteria in water are infectious for human and other hosts and, in the case of the pathogens, pose health risks, remains uncertain and is quite controversial (Bogosian and Bourneuf, 2001; Bogosian et al., 1998; Kell et al., 1998). Some studies have reported that bacteria in the VBNC state have the ability to infect humans or animals (Colwell et al., 1996; Jones et al., 1991). Other investigators have not been able to infect animal hosts with so-called VBNC bacteria or have reported evidence that a few culturable bacteria within a large population of non-culturable bacteria could be responsible for the observed infections (Hald et al., 1991; Medema et al., 1992;
Smith et al., 2002). Because of the lack of scientific agreement of the public health significance of VBNC bacteria and the objections of some authorities even to the use of this terminology, this report does not attempt to address the VBNC issue in the context of microbial indicators of pathogens and human health risks from waterborne pathogens. However, the report does address issues related to the detection of stressed, injured, and otherwise physiologically compromised bacteria in water, the roles and appropriateness of both culture and non-culture methods to detect and quantify bacteria and other waterborne microbes, and the quantitative relationships between bacteria concentrations in water and the human health effects from exposure to water by ingestion and other routes.
The advent of increasingly sophisticated and powerful molecular biology techniques provide new opportunities and alternative approaches to improve upon present indicators and pathogens by both culture and non-culture methods. Molecular methods do not require incubation to culture bacteria because they can directly quantify existing cellular or subcellular structural properties. Therefore, these methods have the potential to be more rapid than culture methods, providing results in as little as minutes to a few hours rather than the typical overnight incubation time for culture methods. Some of these nucleic acid-based methods employ amplification schemes in which a small amount of indicator genetic material is replicated up to a billionfold for easy detection. They also have the potential to be less expensive, making direct measurement of pathogens more economically feasible. Much of the rest of this chapter is devoted to describing the types of molecular methods that are presently under development and have the potential to replace, supplement, or greatly improve the quality of information of classical (largely bacterial) culture-based methods in the future. It is important to mention that Appendix C (Detection Technologies) supplements the discussion (both generally and specifically) of these and other methods by describing them in more detail. Furthermore, molecular methods can be coupled with or linked to microbial culture methods in ways that can increase sensitivity, decrease detection time, and provide conclusive and rapid confirmation of identity and infectivity (e.g., Reynolds et al., 1996).
Targets and Opportunities
Several different analytes can be measured in microorganisms. For purposes of this discussion, microbes can be divided broadly into cells and viruses. Cells can be detected by the following categories of analytes, as summarized in Figure 5-2.
Nucleic Acids
Deoxyribonucleic acids and ribonucleic acids have unique sequences of nucleotide bases (adenine, thymidine [uracil in RNA], cytosine, and guanine) that
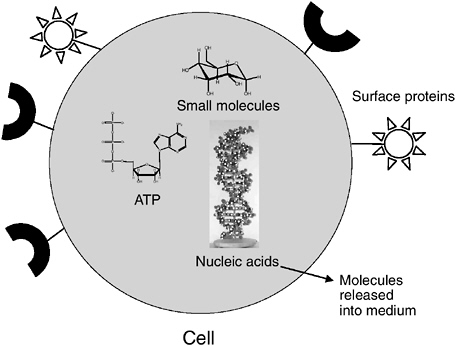
FIGURE 5-2 Targets to measure on or in a cell. Note: ATP = adenosine 5′-triphosphate.
enable the unequivocal identification of a particular organism. DNA and/or RNA is present in all cells and viruses. Cells contain both DNA and RNA, whereas viruses contain either DNA or RNA but never both. The choice of nucleic acids and the ways in which they are measured in microorganisms can provide different kinds of information with regard to microbial identification, viability, and infectivity or culturability. For example, some nucleic acid targets and the methods for their detection can provide very broad identification of a family or genus of microorganism, while other targets can provide very specific identification of species, strain, or subtype. Some nucleic acid targets can be taken as measures of viability or infectivity, such as messenger RNA (mRNA) of cellular microbes or mRNA production by viruses in infected cells. In some cases, mRNA targets are evidence of culturability or infectivity. In general, RNA correlates with viability because nucleases present in most biological samples destroy RNA rapidly. Therefore, both the presence and quality of RNA and the specific sequences present can provide a reasonable indication of viability (see more below). In developing methods to detect and quantify waterborne microorganisms and microbial indicators of pathogens, it is important to consider both the targets for detection and methods of detection with consideration of the value and interpretation of resulting data.
Surface Proteins
Proteins present on the surface of a microbe and to a lesser extent those located within a microbe are often unique and offer a means to definitively identify a microorganism of interest. The most common method of analyzing such proteins is the use of immunoassays in which specific antibodies are raised against the proteins and used as binding reagents. Both monoclonal and polyclonal antibodies can be used. Polyclonal antibodies tend to be more broadly reactive, which makes them useful in detecting microbes as broad groups, such as genera. Monoclonal antibodies have greater specificity because they recognize and bind to a very specific epitope or functional group on or in the target microorganisms. The uniqueness of the epitope depends on its function within the microbe. Some epitopes are common to all members of a microorganism family, genus, or species (group or “common” antigens); others can be highly specific, appearing only in an individual strain, subtype, or variant.
Other approaches to microbe identification based on proteins can employ non-antibody ligands, such as aptamers or phage display libraries, that will specifically recognize and bind to a particular protein or an epitope on it (Breaker, 2002). Such ligand binding probes to identify microorganisms, including bacterial spores (e.g., Bacillus anthracis; Zhen et al., 2002), are becoming more accessible because of the advances made in protein identification and mapping within microbes and the advances made in the synthesis of in vitro proteins, oligonucleotides, or oligopeptides. Certain proteins on the surface or in the interior of microbes can be detected by ligand binding assays. The presence of these markers on or in the cell can be evidence of microbe viability or infectivity. Certain proteins in cells and viruses may be present in a native state only when the microbe is intact and infectious. Therefore, the ability to specifically detect that molecule by a ligand-binding assay can be taken as a measure of viability or infectivity.
Carbohydrates (Polysaccharides)
Carbohydrates or polysaccharides present on the surface of a microbe or within a microbe also can offer a unique way to definitively identify a microorganism of interest. Many of these specific carbohydrates are oligosaccharides covalently bound to proteins to create glycoproteins. Such molecules on the surfaces of cells and viruses often have high specificity or uniqueness in identifying a microorganism. Immunoassays can be used to detect, identify, and quantify such polysaccharides or glycoproteins, again using specific polyclonal or monoclonal antibodies raised against the microbe or the specific target molecules. Like proteins, the specificity of polysaccharide epitopes depends on their function within the microbe, with some antigens common to all members of a microorganism family, genus, or species and others being highly specific for individual strains, subtypes, or variants. Non-antibody ligands also can be used to detect,
identify, and quantify specific polysaccharide epitopes. As with microbial proteins, ligand-binding probes to microbial polysaccharides are becoming more accessible because of the advances made in functional polysaccharide identification and mapping and the advances made in ligand-binding chemistry. As is the case for certain nucleic acids and proteins, the detection of certain polysaccharides on or in microbes by ligand binding can be evidence of a microorganism’s viability or infectivity (Feng and Woo, 2001). Certain polysaccharides in cells and viruses are active receptors for attachment and infection and are present in the native state only when the microbe is intact and infectious. Therefore, detecting such molecules by a ligand-binding assay is a measure of viability or infectivity.
Other Small Molecules
Some microorganisms contain or release characteristic metabolites or products, such as sugars, polysaccharides, antibiotics, alkaloids, lipids, and (protein-based) enzymes and toxins into their environment or growth medium. These compounds may be products of either primary or secondary metabolism and can provide a distinct signature for the microorganism of interest. Many methods are available for analyzing such compounds including mass spectrometry, colorimetric assays, enzymatic assays, and various chromatographic methods. For example, adenosine 5′-triphosphate (ATP) is often measured as an indicator of viable and possibly infectious cells, because it is degraded rapidly when the cell dies (e.g., bioluminescence assays; Deininger and Lee, 2001).
Special Considerations for Viruses
Viruses are typically detected either by their DNA or RNA (for RNA viruses) and their surface proteins (either the capsid or the envelope; see Figure 5-3). Although many viruses do not contain small molecules or detectable amounts of internal protein, most animal viruses do. When present, these internal proteins can also be targets for detection, although often they are less accessible than surface proteins. Because viruses are inert outside their host cells, determining the infectivity of a virus often depends on culturing it in host cells. When they do infect host cells, viruses begin to produce new, virus-specific molecules that can be targeted for detection by molecular and other chemical methods as evidence of their presence, infectivity, and concentration. Virus-specific nucleic acids, such as mRNA and proteins, including both structural and nonstructural proteins, can be targeted for detection by chemical, immunochemical, and molecular methods. In addition, all viruses have specific functional groups or epitopes on their surfaces that are used for attachment to host cells. If the cell receptor or its functional ligand constituent can be identified, such a molecule can be used to detect and quantify viruses through a ligand-binding assay.
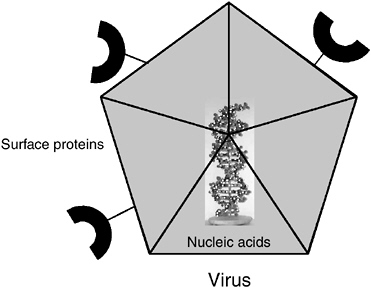
FIGURE 5-3 Targets to measure on or in a virus.
Special Consideration for Protozoa
Like bacteria, waterborne protozoa are single-celled organisms and consist of many of the same components. Unique to the enteric protozoa, however, is the formation of an (oo)cyst as part of its environmental and infectious stage (see Chapter 3 for further information). In most cases, this structure is currently detected by microscopy through the aid of stains and antibodies against the (oo)cyst cell wall. Enteric protozoa are obligate parasites and are similar to viruses in that they need a host organism to reproduce. Thus, determination of the potential viability of protozoa has been studied using vital dye inclusion-exclusion as a measure of the integrity of an (oo)cyst’s outer wall as well as its inner cytoplasmic and nuclear membranes. Huffman et al. (2000) showed, however, that vital dyes grossly overpredicted infectivity of Cryptosporidium under some circumstances. Cell culture methods are now being used and have been found to be statistically comparable to animal infectivity for the determination of infectious oocysts (Slifko et al., 2002). Methods continue to evolve, and as with other microorganisms, polymerase chain reaction (PCR) techniques to target the nucleic acid components as well as methods that combine cell culture and PCR are now being used for detecting protozoa in water (Quintero-Betancourt et al., 2002; see also Appendix C). For example, the free-living amoeba (e.g., Naegleria) can be isolated from water using a culture technique (i.e., their growth in the trophozoite
stage is responsive to a bacterial culture). In addition, PCR, probes, and culture methods are now being combined to identify those species and subtypes that are particularly lethal to humans (Kilvington and Beeching, 1995).
ISSUES IN SAMPLING AND ANALYSIS
The process of making a measurement consists of the four steps shown in Figure 5-4. A common misunderstanding is that measurement is the only critical step in the analysis process.
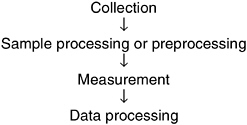
FIGURE 5-4 Four steps involved in performing a measurement.
However, as discussed below, all four components of the process must be considered to ensure accurate analysis of microbial water quality.
Collection and Sampling Issues
The first step in performing a measurement is collecting a sufficiently representative sample, and this remains one of the most challenging problems in water quality monitoring. By representative, it is meant that the sample will reliably portray the presence and concentrations of the analyte of interest (e.g., a microorganism or a chemical) in the water being evaluated or analyzed for its quality. Furthermore, it is important that the sample also be representative of human exposures that may lead to pathogen ingestion and any resulting infection and illness. As noted previously, it is important to recognize that the presence and concentrations of microorganisms and chemicals in water and other environmental media can be highly variable over time (at different times) and space (at different locations within the same body of water). Therefore, as described in Chapter 4, obtaining representative samples often requires taking multiple samples over an extended period (e.g., daily, weekly, monthly), sometimes from different locations within a body of water during the same time period. The importance of addressing variability in microorganism concentrations in water as related to human exposures to pathogens has been well documented in recreational water epidemiologic studies (Fleisher et al., 1993; Kay et al., 1994). The temporal variabil-
ity of microbial occurrence in groundwater has also been documented (EPA, 2000).
Collecting representative samples requires careful consideration of the objectives or purpose of sampling in the context of the need to obtain a reliable estimate of microbial exposure in a timely fashion. Unfortunately, sample collection often involves simply “grabbing” a volume of water and placing it in a storage vessel. For many samples, it is important to preserve the sample, by refrigeration or chemical preservatives, to avoid degradation. All or a fraction of the sample is then taken to the analysis site for further processing. Typically, a sufficient sample volume is taken either to determine whether a microorganism or other analyte is present (i.e., presence-absence) or to estimate the concentration of microbes or other analytes in the water being analyzed (e.g., number of microbes per unit volume).
For microorganisms of public health concern in water, both types of analysis (presence-absence and concentrations estimates) are now used for estimating exposures and making decisions regarding the acceptability of the water for beneficial use under the Clean Water Act (CWA; see also Chapter 1), such as drinking water supply. In some cases, the goal of the analysis is to document that samples of a certain volume (e.g., 100 mL) do not contain a particular microorganism the vast majority of the time (e.g., absence of total coliforms in 95 percent of successive 100-mL drinking water samples) or ever (e.g., absence of Escherichia coli in successive 100-mL volumes of drinking water all of the time). In other cases, the goal of the analysis is to document that samples of a certain volume contain a particular microorganism at concentrations below a threshold level considered indicative of an unacceptable health risk (e.g., maximum allowable concentrations of fecal indicator bacteria in recreational bathing waters). In water analysis based on either presence-absence or estimates of concentration, the variability of microbial concentrations is typically addressed by taking repeated samples from the body of water over time and determining both central tendency (e.g., mean or median) and dispersion (e.g., minimum-maximum values, interquartile range, 95 percent confidence limits).
The focus of data analysis and interpretation is often on typical exposures that are portrayed by central tendencies and dampened extremes, such as 95 percent confidence limits, that are based on logarithmically transformed data. Recent evidence from food microbiology and foodborne disease outbreaks indicates that measures of central tendency and the use of logarithmic transformations of microbial concentration data for the purposes of calculating geometric means and corresponding logarithmic measures of dispersions may be inappropriate for extrapolating to higher exposures and estimating corresponding health risks (Paoli, 2002). Such transformations tend to suppress the effects of extreme values, including the high values on the upper end of a frequency distribution that represent the greatest levels of exposure and health risk. Characterizing the extremes of exposure is necessary because illnesses can result from combinations of rare
events that lead to high levels of exposure. Therefore, data on the magnitude and probability of deviations at the high extremes are needed and must be taken into consideration. The widespread use of logarithmic transformations and measures of central tendency and dispersion of log-transformed data to estimate exposures and health risks needs to be reconsidered in water microbiology, epidemiology, and health risk assessment.
An important characteristic of sampling when there is the likelihood of only low-level detection is that, although the species (microorganism or chemical) of interest may be present in the water being sampled, it may be present at such a low concentration that a given (typical) sample will not contain it. In such situations, the term “Poissonian sampling” comes into play. Simply put, Poissonian sampling aims to determine how many samples of a given volume, or what volume of sample, must be analyzed to ensure that the species of interest is present or not at the prescribed threshold level.
Given a random sample, Figure 5-5 below illustrates the typical numbers involved. The key parameter is s, which is essentially the “average” number of target microbes or molecules that one finds in the sample volume being analyzed. For example, if there are 100 target microbes or DNA genomes per milliliter in a sample, then a sample volume of 10 μL will, “on average,” contain one microbe or genome per sample volume. For lower concentrations of targets, the volume required to ensure a representative sample increases accordingly.
A variety of factors also must be considered in devising and using sample plans when estimating microbial concentrations in water. Sampling may be intended to observe long-term trends in the concentration of microbes in a body of water in which the emphasis is on determining if a measure of central tendency, such as a geometric mean concentration based on replicate samples over a monthly period, is below a specified value. In this situation, the emphasis is on estimating the typical concentrations of microorganisms in water over a long
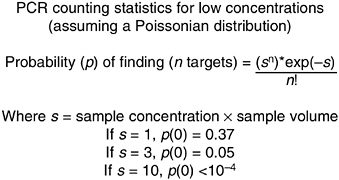
FIGURE 5-5 PCR statistics for low concentrations. SOURCE: Raymond Mariella, Lawrence Livermore National Laboratories, personal communication, 2002.
period of time as a measure of the acceptability of the water for a beneficial use. In such applications, microbial sampling plans still have to address sources of variability in microbial concentrations and the detection of extreme high concentrations of microorganisms from events such as precipitation and other increases in microbial loads. Additionally or alternatively, sampling plans may be intended to estimate the concentration of microorganisms at a single point in time when populations are exposed (e.g., swimmers on a particular day at a bathing beach). In this case, emphasis must be placed on obtaining temporally and spatially representative samples of water to determine whether the concentration of microbes is below that producing an acceptable health risk from a single exposure event (see Chapter 4 for further information). Sampling plans and procedures to estimate the risks from such short-term exposures may have to be quite different from sampling plans and procedures intended to estimate long-term trends and typical concentrations (see also Figure 5-1).
Whether sampling is intended to estimate concentrations for determination of immediate or short-term exposure risks or longer-term trends, it is clear that little information can be obtained from analysis of a single sample of water for a microbial indicator or pathogen. Statistically based sampling methods must be used in conjunction with analyses of multiple samples in order to estimate how microbial levels and human exposures change with varying water quality conditions. Sampling plans must be able to identify when and where microbial concentrations in water are at their highest levels since this is when human health risks are greatest.
Because of the issues associated with collecting representative samples, additional research to develop improved methods for rapid sample concentration and effective, reproducible sample recovery should be supported.
Preprocessing
Once a sample has been collected, it may be subjected to several steps designed to prepare it for analysis of the target microorganisms. For example, in the case of bacterial analysis by culture, a water sample may be filtered to collect the bacteria on a membrane filter that is then placed on a culture medium for incubation and the development of bacterial colonies (see also Appendix C). In this case, the bacteria are separated and concentrated from the sample water prior to culture and enumeration. Similarly, the physical preprocessing steps of filtration or sedimentation by centrifugation have been used to recover microbes from water samples, while at the same time both concentrating them and separating them from other constituents. Several preprocessing steps are available to help purify the sample so that the desired components can be measured without potential interference from other sample constituents. For example, for analysis of nucleic acids it is necessary to remove organic matter (e.g., humic acids) and cellular
debris and metals (e.g., iron, aluminum, heavy metals) because they can inhibit the reactions employed for analysis (Kreader, 1996; Reynolds et al., 1997).
Preprocessing to separate microorganisms or molecular targets in microorganisms from matrix constituents includes chemical methods such as precipitation (with inorganic salts, polyethylene glycol, or acids); solvent extraction (e.g., chloroform); adsorption (to charged surfaces of filters, minerals, or synthetic polymers); chelation (of heavy metals and multivalent cations); chromatography (ion exchange and size exclusion); multiple aqueous phase separation using soluble polymers; treatment with detergents; and ligand binding (e.g., immunocapture, immunomagnetic separation).
A second aspect of processing or preprocessing in biological measurement often involves amplifying the desired microbe or other target analytical component. In some cases, amplification of the target microbe is an essential feature of the measurement method, such as culturing bacteria in liquid (broth) or on solid (agar or membrane filter) media for their quantification or enumeration. Another increasingly used example of processing or preprocessing is PCR, which amplifies a specific DNA sequence that may be present in a sample. More specifically, the DNA sequence of interest is amplified 2n where n = number of PCR cycles. In this manner, the DNA sequence of interest is amplified exponentially, and the resulting sample contains a high concentration of the sequence of interest and can be measured and detected easily. RNA targets, such as the genomic RNA of enteric viruses and mRNA, also can be amplified as a processing or preprocessing method. Usually this amplification is done first by synthesizing a complementary nucleic acid strand (cDNA) to the target RNA sequence by reverse transcription (RT) and then applying PCR to the resulting double-stranded molecule. This method is referred to as RT-PCR.
Quantitative Versus Qualitative Measurements
It is important to recognize that while methods aimed particularly at treated drinking water or groundwater have focused on detecting presence versus absence of a particular indicator in a given volume, in most ambient water applications, obtaining quantitative information has been the ultimate goal. Thus, it is important to obtain reliable estimates of concentrations of target microorganisms or the indicator being measured. The Most Probable Number (MPN) statistical approach has long been used in environmental water microbiology along with quantal (i.e., presence-absence) assays and has been automated to the extent that labor associated with the dilutions and replicate assays is now less tedious and costly (e.g., semiautomated quantification, liquid-based methods for E. coli such as Quanti-Tray®). Furthermore, to establish or study risk estimates associated with a given water sample, the concentration of the pathogen or indicator may be required. Threshold concentrations of certain pathogens or indicators must often be determined to assess whether the water is in compliance with regulations. A
simple presence-absence measurement without quantification is insufficient in such cases. In other cases, such as the deliberate introduction of a toxic chemical or biological threat agent nominally not found in water, a qualitative presence-absence determination is generally sufficient because it indicates that a problem exists. As noted previously, it is important that quantitative measurements consider extreme events (high concentrations) and reliably represent the frequency distribution of these events and their temporal and spatial variability.
Measurement
Once a sample has been suitably processed or preprocessed, a measurement is made. This step may involve injecting the sample into an instrument; titrating a sample; scoring cultures as positive or negative for microbial growth; or enumerating colonies, plaques, or foci present on agar or membrane filters or in infected cells after a requisite incubation period. The end result is to collect data in the form of spectra, counts, volume, optical density, and so on. These data are simply values that correspond to some parameter being detected by the instrument or by the individual taking the readings. Various methodologies widely employed for making measurements of microorganisms in water samples are summarized later in this chapter and in Appendix C.
Data Collection and Processing
Once measurement data have been scored, they are collected (compiled) and processed. Processing involves manipulating or analyzing the data based on the presence, absence, or concentration of the analyte being analyzed. In simple cases, data processing is straightforward—for example, scoring the presence or absence of a particular analyte (e.g., virus or bacterium) by simply observing and recording the positive or negative result obtained during the measurement phase of analysis. In other cases—for example, estimating the concentrations of microbes cultured in different and replicate volumes of broth media—the numbers of positive and negative cultures of the total cultures inoculated per sample volume have to be processed through calculation of an MPN or a 50 percent infectious dose (ID50). Some measurements generate complex or large amounts of data that must be subjected to detailed analysis before a result for the presence, absence, or concentration of the target analyte can be determined. For example, amplified nucleic acid from PCR may have to be subjected to nucleic acid hybridization or nucleotide sequencing before the sample can be confirmed as positive. Mass spectral data must be processed to correlate the measured spectrum to the spectra of various compounds stored in a database. With the increasing amount of data coming from high-density arrays, mass spectra, and long-term time series with high spatial coverage, there is a need to devote additional resources and effort to data storage and processing.
ASSESSMENT OF METHODS AND THEIR ATTRIBUTES
In analyzing water and other environmental samples for indicators of pathogens or for pathogens themselves, three main options are available: (1) analyze for live or infectious microorganisms (pathogens or indicators); (2) analyze for microorganisms without conclusively determining their infectivity or viability; or (3) analyze for another constituent in the sample (a surrogate) that is indicative and predictive of the presence and concentration of the pathogen or microbial indicator (e.g., a chemical associated with fecal contamination; see Chapter 4 for further information).
Direct Analysis of Microorganisms by Infectivity, Culturability, or Viability
On the basis of indicating public health risk of infection or disease from exposure to microbially contaminated water, the direct measurement of a pathogen or a reliable microbial indicator of pathogens by culture or infectivity is generally considered the “gold standard,” and both should be the goal of any new measurement technique. That is, if a pathogenic microorganism can be cultured and shown to be infectious, it indicates that the organism is viable and potentially able to cause infection and disease given sufficient exposure and a susceptible host. Analyzing for a microbial indicator by culture or infectivity can also be predictive of such a health risk, provided the indicator is otherwise a reliable predictor of infectious pathogens. Various methods of analyzing for pathogens by culture or infectivity are available and have been reviewed and described in detail elsewhere (Hurst et al., 2002; Sobsey, 1999, 2001).
Briefly, the most commonly used culture methods for bacteria are colony counts on membranes or agar medium plates and liquid broth cultures. In either format for culture, the target bacteria are detected by and distinguished from other bacteria by use of differential and selective media that have specific ingredients for multiplication of the target bacteria, such as chemical inhibitors against the growth of non-target bacteria, and constituents (specific growth substrates or indicator chemicals such as oxidizing or reducing agents) that cause the growth of the target bacteria to be unique in appearance and distinguishable from non-target bacteria (differential ingredients).
Because viruses are obligate intracellular parasites, culturing them requires the use of susceptible host cells. The viruses will attach to and penetrate the host cell, where they will multiply (replicate), usually with subsequent release of progeny viruses and death and lysis of the host cell. This process of infection can be detected by death or lysis, as manifested by the disappearance of the cells (“clearing”) from a broth culture; the development of virus-induced morphological changes in the appearance of the cell (cytopathogenic effects, or CPEs); or the development of discrete circular, cleared areas in a layer or lawn of cells in an
agar medium, which are referred to as plaques. Some viruses do not produce visible lysis or CPEs in host cells although the viruses have multiplied within the cells. In such cases, the presence of the viruses in the infected cells can be detected by molecular methods (nucleic acid, protein, or enzyme) or by immunochemical and immunohistochemical methods, as described later in this chapter.
The application of cell culture for Cryptosporidium parvum infectivity began in the early 1990s and was reviewed recently by Rose et al. (2002). A variety of end points are currently being utilized to determine the concentration of infectious oocysts in water samples. Immuno-based assays utilizing antibodies to C. parvum sporozoites and other life-cycle stages, coupled with a secondary antibody conjugated to a fluorescent dye or enzyme, have been employed. Molecular-based assays using either PCR or RT-PCR methodologies to amplify DNA or RNA targets extracted from infected cells or oligonucleotide probes that can detect nucleic acids in situ have also been developed for speciation and genotyping. In a study of surface waters and filter backwash waters, infectious oocysts of a variety of strains were detected in 4.9 and 7.4 percent of the samples, respectively, using cell culture methods (DiGiovanni et al., 1999).
Another way to analyze for pathogens or microbial indicators of pathogens is by direct observation of their viability. Viability can be analyzed on the basis of several different measures or end points, such as physical movement (e.g., of larvae in ova), hatching (e.g., excystation of protozoan cysts or oocysts), enzyme activity, oxidation-reduction, synthesis of macromolecules, and uptake or exclusion of dyes. In the case of some microorganisms, viability measurements are likely to be good predictors of infectivity because the end point is actually detecting the activity of a living organism. For example, the detection of viable helminth ova by microscopic examination for the movement of mature larvae within the ova is likely to be strongly associated with human or animal infectivity. However, some viability assays, such a excystation of protozoan (oo)cysts or dye exclusion (or uptake), are poor predictors of infectivity for human or animal hosts.
Analysis of Microorganisms by Measurement of Their Constituents or Components
As noted previously, measuring components of microbes is often used for their analysis in water and other environmental media, and some of these approaches are summarized below (for further information see Hurst et al., 2002; Sobsey, 1999, 2001). Although the techniques described below are designed to measure specific components of the microorganism as discussed elsewhere in this report (see also Box 4-2), the most important biological attribute of an indicator is a strong quantitative relationship between indicator concentration and the degree of public health risk. That analysis of the constituent should itself be a
reliable predictor of human health risk from exposure to microbiologically contaminated water.
Strategies
For all the non-culture based methods of microbial analysis, there are two general approaches that can be used to identify the presence of a specific microorganism: (1) targeting a single specific component of the organism that is unique and characteristic of that organism and (2) using fingerprinting in which a pattern of components signifies the presence of the microorganism. In the latter case, the individual components may not be unique to the particular microorganism of interest, but their concentration, co-occurrence, or sequence generates an overall response profile characteristic of the microorganism.
Nucleic Acid Analysis
DNA sequences can be present at only one copy per cell, which poses an extremely difficult detection challenge. Some target genes, however, such as unique intragenic sequences in DNA and certain forms of RNA, can be present in multiple copies in a cell, making these sequences easier to detect. DNA is typically amplified first using PCR (described earlier and more extensively in Appendix C). PCR is sequence specific, although sometimes in the absence of a specific target sequence, nonspecific amplification of non-target sequences may occur; in addition, under low stringency conditions non-specific binding of the primers also occurs. These nonspecific products will usually give negative results in subsequent analysis of the PCR products. PCR followed by analysis such as hybridization provides two levels of discrimination.
Another method based on hybridization is fluorescent in situ hybridization (FISH). In this approach, fluorescent probes specific to different regions on the chromosome containing different labels are hybridized to the intact microorganism and the pattern of colors on the chromosome are viewed by microscopic examination under a fluorescent microscope. The unique banding pattern corresponds to the microorganism of interest.
RNA is present in bacterial cells or protozoa as messenger RNA (mRNA), ribosomal RNA (rRNA), or transfer RNA (tRNA). In RNA viruses, the RNA is present as the viral genetic material, as either a single- or double-stranded molecule, or as either one continuous strand or multiple unique strands. Messenger RNA is present in many copies per cell and, as described above, is typically first converted into complementary DNA by reverse transcription and then PCR-amplified for analysis (RT-PCR).
Nucleic acid analysis can be conducted using several strategies. Specific unique sequences can be selected that have no counterparts in any other microorganism. In this manner, the presence of the amplified DNA (the amplicon) sig-
nals the occurrence of the specific sequence of interest. In this type of analysis, prior sequence information about the specific organism of interest is required. It is important to note that the uniqueness of nucleic acid sequences can vary—from sequences that are common or shared among closely related microbes and can therefore detect families, genera, or other groups of microbes, to highly specific sequences that can identify a single species, strain, or variant of a microbe. Alternatively, DNA or RNA fingerprinting is performed in which the pattern of nucleic acid sequences is correlated with a particular microorganism. In this case, sequence information is not essential as long as the pattern is known a priori. In this approach, the identifying pattern is based on a reference microbe or microbes. In most environmental applications, a collection of reference microorganisms, called a library, is created against which to compare the pattern observed in an environmental isolate obtained from a sample (see Chapter 4 for further information).
After amplification, DNA is analyzed either by sequencing or by hybridization to a unique complementary genetic sequence (a gene probe) or to an array containing the complementary genetic sequence. For example, Dombek and colleagues (2000) used membrane filtration to first concentrate E. coli samples from a variety of sources (humans, chicken, cows, ducks, geese, pigs, and sheep) and then microarray technology (DNA fingerprinting; see Appendix C for a detailed discussion of microarrays in detecting waterborne and foodborne pathogens) to identify their sources with success rates ranging from 89.5 to 100 percent. If only one or a limited number of sequences are required for identification, rapid or real time PCR can be employed in which a fluorescence signal appears only when the sequence of interest is present. The advantages of rapid (real time) PCR over traditional PCR methods include faster results and fewer handling steps (see Appendix C for further information). Rapid PCR methods are amenable to field use, and several commercial vendors have instruments available for bio-warfare agent field detection. Their use to detect enteric microbes in field samples has already been reported (Donaldson et al., 2002). Therefore, as these methods improve and become more widely available, there is considerable promise for the expanded application of this rapid PCR technology to detect microbial pathogens and indicators in environmental waters.
Although these molecular biology methods directed at nucleic acids were developed primarily for the Human Genome Project1 with application in clinical medicine, these powerful techniques have direct applicability to waterborne pathogen or indicator detection is clearly feasible and has already been done (Cook, 2003; Griffin et al., 2003; Keer and Birch, 2003). So far, much of the application of nucleic acid amplification, detection, and characterization is by
|
1 |
See http://www.ornl.gov/TechResources/Human_Genome/home.html for further information. |
hybridization using macro-scale methods (e.g., various forms of conventional PCR and RT-PCR and filter or other hybridization to detect amplicons). However, microarray technology is becoming increasingly available for use in applied and environmental microbiology, primarily directed at gene expression of living microorganisms under different environmental conditions. In these applications, the identities of the microorganisms may already be known, many cells are exposed to the conditions under study, and numerous copies of the target nucleic acid are available for detection. DNA microarrays have been applied successfully to the detection of pathogenic microorganisms in environmental samples. Direct detection of extracted or accessible nucleic acids is possible when present at high concentrations (>10 cells per mL; Chandler et al., 2001), and for low numbers of target organisms, nucleic acid amplification is applied prior to hybridization (including in microarrays), nucleic acid sequencing, or other detection and characterization technologies.
One of the important issues to address in the application of nucleic acid technologies to the detection of pathogens and indicators in water and other environmental samples is to what extent and how such technologies can distinguish infectious and viable microbes from those that are noninfectious or inactivated, including the detection of nucleic acids from dead and degraded microorganisms. It is important to recognize that the detection of waterborne pathogens or indicators whether viable and potentially infectious or not, may provide sufficient information to assess vulnerability to contamination or to make decisions about public health risk (see also Chapters 4 and 6). For example, under circumstances where the basis of acceptable risk is the absence or not exceeding a specified maximum concentration of the nucleic acid of a pathogen or indicator in a specified sample size. Such a management approach has been proposed for Noroviruses or F+ coliphages as Norovirus indicators in raw bivalve molluskan shellfish (Dore et al., 2000, 2003).
Under some other conditions, the detection of nucleic acids from inactivated microorganisms would not necessarily be indicative of a health risk and would not be considered useful information in support of management decisions. For example, water subjected to physical or chemical disinfection that inactivates viruses and other microbes can still contain detectable nucleic acids of these inactivated viruses (Sobsey et al., 1998). One approach to overcoming the problem of detecting the nucleic acid of inactivated viruses is to couple nucleic acid detection methods such as PCR with microbial culture methods. Initial amplification of the microbes through culturing is then followed by methods to detect their nucleic acids. This has been done successfully for enteric viruses and is referred to as integrated cell culture-PCR (ICCPCR; Blackmer et al., 2000; Reynolds et al., 1996).
Another nucleic acid approach to detecting viable and potentially infectious microorganisms is to target only those nucleic acids found in organisms in this state, such as messenger RNA. For viruses, another approach is to detect only
fully intact and potentially infectious RNA viruses by first exposing them to proteases and then ribonucleases, to which intact and potentially infectious viruses are resistant. These enzyme pre-treatments degrade damaged and noninfectious viruses, leaving only the intact and potentially infectious viruses to be detected by subsequent nucleic acid amplification methods (Nuanualsuwan and Cliver, 2002). Yet another approach to the detection of infectious viruses is to amplify only full length viral genomic nucleic acid. If the nucleic acid has been degraded to less than full length fragments or contains lesions causing inactivation, then nucleic acid amplification does not occur. Thus, several promising methods to the detection of the nucleic acids of only intact, viable, and infectious microorganisms are in development and being validated.
As the methods for recovery, concentration, and purification of target microbes and their nucleic acids are further improved for the application of various nucleic acid methods, including microarray technology, it is likely that these approaches will become more widely applicable to the detection, quantification, and identification of microbes in water and other environmental media. The committee concludes that the introduction of molecular techniques for nucleic acid analysis is a growth opportunity for the field of waterborne pathogen detection and recommends that U.S. Environmental Protection Agency (EPA) resources be invested to accelerate the introduction and further development of these techniques. Lastly, it should be noted that microbial toxins, which are proteins, cannot be detected by PCR or other nucleic acid analysis-based methods.
Immunological Methods
For surface proteins, large peptides, and their glycosylated derivatives, immunological methods offer a high degree of specificity and sensitivity. Although small molecules such as toxins generally cannot be detected using immunological methods, surface proteins on bacteria, protozoa, and viruses can be unique to the microbe and detected by immunological methods of analysis. In this approach, an antibody is raised against the microorganism or the purified protein to be detected. Both polyclonal and monoclonal antibodies can be obtained, and an immunoassay is developed around these antibodies. Typically, the antibodies are employed to capture the analyte, carry a label to the analyte, or both. The most common immunological method used is the enzyme linked immunosorbent assay (ELISA) in which a capture antibody bound to a surface is used to bind and concentrate the analyte. A second antibody, labeled with an enzyme, is then bound to a second recognition site on the analyte. Finally, a chromogenic or fluorogenic substrate is added that is converted to an observable product, which can be detected. A detailed description of the ELISA method appears in Appendix C.
Another immunological method now becoming widely used in environmental microbiology is immunomagnetic separation (e.g., Gehring et al., 1996; Mitchell et al., 1994). This method can be employed as both a processing (recov-
ery, concentration, and purification) method and a detection method (or at least part of a detection method). The typical application of immunomagnetic separation is to have the antibodies bound to a solid phase, such as paramagnetic beads, and then react those beads with the sample for target microbe recovery by its binding to the antibody on the solid phase. Typically, another method, such as culture, immunofluorescence microscopy, or nucleic acid hybridization or amplification, is then used for detection and confirmation of the target microbe or its components. In some applications, reaction of the target microbe antigen with the antibody on the bead is a sufficient basis for detection using an electronic sensor.
For example, an electrochemiluminescence (ECL) technology for detecting Cryptosporidium parvum oocysts in environmental water samples has been recently developed (Lee et al., 2001). The method is reported to be quantitative and reproducible, and requires only minimal sample processing. Currently, the ECL assay detects as few as 1 oocyst in 1 mL of concentrated test sample with sample turbidity of up to 10,000 nephelometric turbidity units (NTUs). In this study, water and sewage samples collected during a cryptosporidiosis outbreak were tested by ECL assay. Cryptosporidium parvum oocysts were found in the source water at the time of outbreak, and a sharply decreasing level of oocysts in sewage samples was observed over a three-month period following the outbreak. The use of immunocapture technologies in conjunction with electrochemical detectors is one of several approaches to rapid and improved immunological detection of waterborne pathogens and indicators.
Another direction for further advancement of immunodetection is for viable waterborne microorganisms. For example, a quantitative immunoassay capable of detecting low numbers of excystable, sporozoite-releasing C. parvum oocysts in turbid water samples has been developed (Call et al., 2001). Monoclonal and polyclonal antibodies have been developed against a sporozoite antigen released only during excystation or when the oocyst is mechanically disrupted. In this assay, oocysts in the test sample are first excysted and then centrifuged. The soluble sporozoite antigen is captured by monoclonal antibodies attached to a magnetic bead. The captured antigen is then detected by ruthenium-labeled polyclonal antibodies via electrochemiluminescence. This viability assay can detect as few as 50 viable oocysts in a 1-mL assay sample with a turbidity as high as 200 NTUs. With further development, refinement, and validation, immunoassays may eventually be able to detect a variety of different viable microbes in water and other environmental samples.
Another immunologically based method is flow cytometry. In this method, microbial cells are labeled with a fluorescently-labeled antibody. Multiple antibodies are employed, with each antibody specific to a particular microorganism. The labeled mixture is then passed through the flow cytometer, which interrogates the solution and determines the numbers of each microorganism based on the occurrence of each label (e.g., Collier and Campbell, 1999; Veal et al., 2000).
Miscellaneous Methods
A wide variety of other available methods can be employed to measure constituents or components of cells. One approach to molecular detection of waterborne pathogens and indicators is based on the use of ligand-binding assays to recover and detect target microbes. Many microbes possess specific surface receptor molecules or epitopes that bind to specific molecular targets and have various functions, such as cell attachment, transport of molecules for nutrition, or molecular processing for immune response or other biological activities. As these molecules and their corresponding targets are elucidated, the molecules to which these epitopes bind can be used for microbe capture and detection—analogous to the use of antibodies for such purposes, as described earlier. Because some microbe surface receptors are used for initiation of infection in host cells, the ability of the microbes to bind to their specific target ligand can be used to detect intact, chemically functional, and potentially infectious or viable microorganisms. Such assays for viability based on the ability to bind to specific receptors are now under development for waterborne microorganisms and are likely to be developed further.
Other chemical and biochemical constituent analysis also can be used to ascertain the presence of viable organisms (e.g., ATP detection with luminescence detection; Deininger and Lee, 2001), the presence of specific toxins, or the organism’s protein profile. One of the more powerful and increasingly used analytical methods is mass spectrometry, which is employed for whole organism analysis as well as small molecule analysis. In the former case, bacteria or bacterial spores can be injected directly into the mass spectrometer and their lipid and/ or protein fragmentation profiles can be used to identify them (Ishida et al., 2002; Madonna et al., 2001; see Appendix C for an example). Alternatively, using preconcentration followed by front-end separation such as gas or liquid chromatography coupled with mass spectrometery detection, small molecular components can be analyzed. Significant advances will be required before mass spectrometry can be used in the field because the instruments are generally large, and require significant amounts of power.
Attributes of Methods
All of the methods described thus far have both positive and negative aspects associated with their use in the detection of waterborne pathogens or microbial indicators of pathogens. Table 5-1 provides a qualitative description, based on the collective expertise of the committee, of how each major grouping of detection methods (i.e., culture, immunological, nucleic acids, cell components) currently performs relative to each desirable method attribute described in Chapter 4 (see Box 4-3). In some cases, there is a wide range of performance for each attribute within a given method.
Multi Parameter Measurements
Microbial methods can be designed to measure a single parameter or multiple parameters to detect and quantify microorganisms. For example, some culture methods detect and quantify microorganisms by the ability to display several parameters such as growth in a lactose medium at 44.5°C with acid and gas production as the basis for fecal coliforms analysis. Other methods are based on the ability of the target microbe to utilize a specific array of organic substrates as a basis for its identification, whereas still others—such as immunological detection with a specific monoclonal antibody or PCR amplification with a specific primer set—may detect only a single organism or closely related group of microorganisms. As noted throughout this report, at present it is impossible at present to completely capture and characterize the microbial quality of water for all pathogens by any of the currently available methods. With new and emerging technologies, it is likely to become possible to simultaneously measure multiple analytes in a water sample, thus providing a better basis for judging the microbial quality of the water from which it is taken and any associated health risks. Although any of these methods can be used for pathogen detection and identification, the simultaneous use of multiple capture methods based on orthogonal detection principles (e.g., antibodies and nucleic acid probes) can significantly increase detection specificity of waterborne pathogens and their indicators.
Three strategies can be used to obtain multiparameter measurements: (1) integrate data from many different measurement technologies, (2) integrate several measurement techniques into a single system, and (3) develop instruments that have the intrinsic ability to make multi-analyte measurements (e.g., arrays). The committee recommends multiparameter approaches in which many technologies and methods are integrated to obtain the best possible information from available samples.
More consideration should also be given to “broad range” survey methods, for example, broad range ribosomal RNA or DNA PCR with high throughput sequencing, DNA microarray-based analysis, or mass spectrometry-based analysis of PCR products. While these approaches are not currently ready for widespread routine use, they are critical for building databases dedicated to background characterization and identification of predictive patterns for waterborne pathogens and indicator organisms.
STANDARDIZATION AND VALIDATION OF METHODS
Whatever indicators or pathogens are ultimately selected as the best suited for measuring microbial water quality to achieve specific applications, several methods are already available or on the horizon. Such methods may include more conventional methods based on cultivation and membrane filtration, perhaps using new formulations of biochemical media, or they may be novel and use inno-
TABLE 5-1 Comparison of Major Categories of Microbial Detection Methods by Desirable Attributes
|
Method Attributes |
Culture |
Immunological |
Nucleic Acid |
Cell Component |
|
Specificity to desired target organism |
Low to moderate |
High |
High |
Moderate to high |
|
Broad applicability |
High |
High |
High |
Low to moderate |
|
Precision |
Moderate |
Low to high |
High |
Moderate to high |
|
Adequate sensitivity |
Moderate to high |
Low to moderate |
High |
Not applicable |
|
Rapidity of results |
Low to moderate |
Moderate to rapid |
Moderate to rapid |
Low to moderate |
|
Quantifiable |
Moderate |
Low |
Low to moderate |
Not applicable |
|
Measures viability or infectivity |
Yes |
No |
No, but possible |
No, but possible |
|
Logistical Feasibility |
||||
|
Training and personnel requirements |
Low to high |
Moderate |
Moderate |
Low to high |
|
Utility in field |
Low to high |
Moderate to high |
Moderate to high |
Low to high |
|
Cost |
Low to high |
Moderate |
Moderate |
Low to high |
|
Volume requirements |
Low to high |
Low |
Low |
Low to high |
vative and emerging technologies such as biosensors based on antibodies or microarrays based on nucleic acid hybridization. Building on these advances, it is possible to divide measurement methods into two broad, but not mutually exclusive, categories:
-
Research methodologies: methods that have been published but are used primarily in academic, industry, and government research laboratories
-
Conventional methodologies: methods that have been assessed with some model of standardization, are widely accepted and used, and are applicable to industry and private laboratories
The data, research, and information needs (both short and long term) to advance “research methods” into those considered “conventional” are central to the following discussion and the committee’s statement of task (see Executive Summary and Box ES-1). In this regard, the ability to provide timely, accurate, and reliable data is central to the goals of water quality monitoring, testing, and reporting. Thus, the process of method development and validation is directly linked to the quality of data. Standardizing and then validating prospective methods can follow several models, but most have in common a prescription and terminology regarding their specified application (see Box 5-1). The approach also focuses on a number of the methods attributes described in Chapter 4 (especially Box 4-3) and Table 5-1. Thus, a guide for those who are examining new methods or modifying existing methods for new applications should include a description and study of these key elements in the published literature to enhance the method’s potential to be improved or to benefit from wider acceptability and use.
Organizations Involved in Developing Standards
Several organizations and associations are involved in the development of standard methods for evaluating water quality or microorganisms in water (see Box 5-2 and Table 5-2). These methods have been developed primarily in response to industry needs and in some cases to the needs of local, state, or even the U.S. government. Although many standards are developed according to a consensus process in which the views of all stakeholders on the scientific basis of the approach are taken into account, many standards are also voluntary and focus on international standardization that is based on voluntary involvement of all interests. In general, the need for a standard is first articulated and then defined by a group of technical experts, the details are negotiated, and there is finally an approval by the organization and its members. It is important to note that the defined technical aspect for a standard may or may not involve testing and the explicit consideration of the attributes of a method through a validation process.
For the purposes of this report, the primary groups involved in standard development for the microbiological assessment of water include the EPA, Interna-
|
BOX 5-1 Standards involve the development of a common language for something established by an authority or by general consent that can be established and used for the measure of quantity, weight, extent, value, or quality; they are a means for determining what a thing or process should be. Standardization in the field of water quality includes definition of terms, sampling of waters, measurement, and reporting of water characteristics. Validation is the process of demonstrating that a method is acceptable for its intended purpose. The accuracy of a measurement is defined by how close a result comes to the true value and determining the accuracy often involves calibration of the analytical method with a known standard. Precision is a measure of the reproducibility of measurements and is usually described by the standard deviation, standard error, or confidence interval. Precision relates to the quality of an operation by which a result is obtained. Specificity is a measure of the ability of a method to discriminate the desired target accurately (e.g., the microorganism, protein, genetic sequence) in the presence of all potential sample components, including other microorganisms. The response in test mixtures is compared with the response in solution containing only the target. The sensitivity or detection limit of a method is the lowest concentration that produces a response detectable above background or noise level of the system. |
tional Organization for Standardization (ISO), American Public Health Association (APHA), American Water Works Association (AWWA), Water Environment Federation (WEF), and American Society for Testing and Materials International (ASTM).
As noted throughout this report, EPA currently has published methods for various bacterial, protozoan, and viral indicators and pathogens in response to specific rules and programs under the Safe Drinking Water Act (SDWA) and the CWA. EPA’s current validation process for microbiological method is discussed
|
BOX 5-2 In the United States, many organizations comprise the U.S. standardization system; these include government and nongovernmental organizations involved in the development of both mandatory and voluntary (consensus) standards. Mandatory standards are set by the government, and regulatory standards focus generally on health, safety, environmental, or other criteria. Regulatory agencies such as the EPA regularly reference hundreds of existing voluntary consensus standards, in lieu of developing their own, that have the force of law once they are referenced in a government regulation. In this regard, the U.S. National Technology Transfer and Advancement Act of 1996 (Public Law 104-113) requires federal agencies to adopt private sector standards, particularly those of standards-developing organizations (see also Table 5-2), wherever possible rather than creating proprietary, nonconsensus standards (see http://ts.nist.gov/ts/htdocs/210/nttaa/nttaa.htm or http://astm.org for further information). |
later. The ISO methods that are relevant to this report are found under TC 147/SC 4 (TC refers to the technical committee; SC refers to the subcommittee) that developed the microbiological methods. There are 19 published ISO standards including standards for coliforms, Escherichia coli, Clostridium, bacteriophage, Salmonella, Pseudomonas (fluorescens and aeruginosa), Legionella, Campylobacter, Cryptosporidium, and Giardia.
The APHA, AWWA, and WEF regularly publish Standard Methods for the Examination of Water and Wastewater, which is currently in its twentieth edition (APHA, 1998) and remains one of the oldest publications used worldwide for water testing methods. The current edition includes more than 400 methods detailed in a step-by-step format; each method describes the applications and potential uncertainties associated with its use. The work of identification, selection, and ultimate inclusion of prospective methods is conducted by volunteer review committees that utilize the published literature to produce a consensus-based standard method. At present, there are standard methods for bacteria, enteric viruses, and enteric protozoa; however, only the coliform bacteria, E. coli, and heterotrophic bacteria methods are approved by EPA for use.
To date, the standardization of microbiological methods in the United States has generally followed one or more models, but most have in common consideration of the intended application, and one typical route to standardization is inclusion in the aforementioned Standard Methods for the Examination of Water and Wastewater. Another typical route to the standardization of a microbiological
TABLE 5-2 Select Organizations Associated with Standards Development
|
Organization(s) and URL |
Type |
|
American National Standards Institute (ANSI; www.ansi.org) |
Not-for-profit, nongovernmental organization |
|
Association of Analytical Communities (AOAC International; www.aoac.org) |
Not-for-profit organization with ties to federal government funding |
|
American Society for Testing and Materials International (ASTM; www.astm.org) |
Not-for-profit organization |
|
American Public Health Association (APHA; www.apha.org); American Water Works Association (www.awwa.org); Water Environment Federation (WEF; http://www.wef.org) |
Not-for-profit health, drinking water, and wastewater associations (respectively) |
|
U.S. Environmental Protection Agency (www.epa.gov) |
U.S. government regulatory agency |
|
International Organization for Standardization (ISO; www.iso.ch) |
Private agency headquartered in Geneva, Switzerland |
|
National Institute of Standards and Technology (NIST; www.nist.gov) |
U.S. government agency |
|
aMultilaboratory testing entails the evaluation of a method with a highly specified protocol using multiple tests to evaluate inter-laboratory (generally 5 to 11 laboratories) precision and accuracy of the method that requires coordination of reagents and matrix spikes. |
|
method is through the D-19 ASTM group for water. To be published, each method must follow a prescribed interlaboratory testing protocol, with 11 laboratories participating, defining accuracy and precision. Similar to publication in Standard Methods, the application for each method evaluated is described to include the specification of limitations. ASTM has developed microbiological methods for water, including coliphage, enteric protozoa, enteric viruses, and heterotrophic bacteria. EPA had worked closely with ASTM and provided appreciable funding
|
Role and Activities |
|
Does not develop standards; serves as the U.S. member body to the ISO in the development of ISO standards; accredits standards-developing organizations according to their consensus processes and accredits standards developed by others as American National Standards |
|
Previously known as the Association of Official Analytical Chemists. An international provider and facilitator in the development, use, and harmonization of validated analytical methods and laboratory quality assurance programs and services; AOAC provides three methods validation programs (described elsewhere) that require multilaboratory assessmenta |
|
Develops and publishes voluntary consensus standards for materials, products, systems, and services nationally and internationally that require multilaboratory assessment |
|
Develops committees and editorial board for the publication of Standard Methods for the Examination of Water and Wastewater; its 20th edition (APHA, 1998) is approved for use by the EPA |
|
EPA’s Office of Science and Technology is responsible for preparing standards to be used in support of government regulations. EPA publishes laboratory analytical methods that are used by industrial and municipal facilities in analyzing the chemical and biological components of wastewater, drinking water, and other environmental samples required by EPA regulations under the authority of the CWA and SDWA (see also Tables 1-1 and 1-2). Almost all such standards are published by EPA as regulations under Title 40 of the Code of Federal Regulations (CFR) and require multilaboratory assessment |
|
Develops voluntary standards; its membership is comprised of the recognized national standard setting bodies of 140 nations. ISO has more than 180 technical committees devoted to almost all areas of standardization. Final publication of an ISO standard requires the majority consensus of technical committee members and two-thirds of the ISO voting membership |
|
Assists U.S. industry in the development and application of technology, with leading expertise in the area of technology standards and industry standardization issues; is also actively involved in voluntary consensus standards development activities |
for interlaboratory testing; however, no major funding has been provided for this mechanism in the last 10 years.
Validation Process
The development of a standard method and implementation for a biological measurement through validation can be a long, tedious, and expensive proposi-
tion. The validation process itself, however, can focus on assessing the method attributes in a systematic manner. Thus, as certain attributes of a method are evaluated and ranked as most important for a given application, this can drive the validation process as well as the criteria that are established for acceptability. Although methods used for many regulatory actions must undergo more rigorous testing, those used for gathering information that may be employed in more adaptive management strategies might not have to meet such stringent criteria.
Validation of a measurement method includes studies on specificity, linearity, accuracy, precision, range, detection limit (sensitivity), and consideration of robustness and related issues. Early stages may focus primarily on specificity, linearity, accuracy, and precision; however, the validation process should be considered iterative, particularly as a variety of water matrices are tested.
Specificity Studies
Whether using whole cell, cultivation, genetic, or antibody methods, the detection of the specific target will have to be assessed against a variety of other targets. Some methods may try to capture a broad group of organisms (e.g., all enteroviruses, whether animal or human); others may focus more narrowly (e.g., coxsackieviruses from humans only). The specificity is described as the ability of the method to correctly classify organisms (in groups or as specific species or subtypes).
Linearity Studies
Linearity can be evaluated by preparing standard solutions at various concentration levels (five to six have generally been recommended), thus demonstrating the performance of a method over a range of organism densities. Interestingly, nucleic acid-based methods generally have been designed and tested with varying concentrations after extraction (i.e., using dissolved DNA or RNA as the target). It is recommended that new methods also be tested using whole microorganisms, rather than just extracted DNA or RNA targets for linearity and specificity studies.
Accuracy Studies
There are generally three approaches for determining the accuracy of a biological measurement method. First, a sample with a known concentration is tested; however, reference standards for microorganisms may often be difficult to obtain so this approach is not used very often. Second, and more commonly, the new method is compared to an existing method that is accepted as accurate. This approach makes it difficult, however, to test a new molecular method against an
accepted cultivation method because they are measuring different targets. The third approach focuses on calculating the recovery of known numbers of a microorganism spiked into a water matrix. Both blank matrix spikes and various types of water are typically used. Triplicate tests at a minimum of three levels over a range of 10 to 1,000 times the target concentration are often used. As discussed previously, measuring aquatic microorganisms may involve several steps or a series of methods added together for concentration, purification, and detection. Thus, a series of recovery experiments may be required at each stage of the process, or it may prove more expeditious to evaluate the entire method. The range and detection limits of a method are determined from the linearity and accuracy studies.
Precision Studies
There are three levels to the evaluation of precision: (1) intralaboratory tests performed by one individual, (2) intralaboratory tests performed by multiple analysts, and finally (3) tests performed through interlaboratory testing (i.e., multiple analysts from multiple laboratories). The precision of a method can then be determined through analysis of the amount of scatter in the results obtained from multiple analyses of a homogeneous sample. The exact sample and standard preparation procedures that will be used in the final method should be tested. Statistical equivalence is often used to evaluate the results from different laboratories, or the range of results is used to evaluate acceptability.
Robustness and Other Considerations
It is important to note that interferences in the use of a method may affect the results because of constituents in the water matrix (e.g., concentration of organic material, pH). Thus, the sensitivity to such variables helps define the robustness of a prospective method or its ability to remain unaffected by changes. In addition, maintenance of instruments, stability of reagents, and types of controls to be used will have to be described in the validation of a microbiological method.
The level of false positives and false negatives associated with the use of a method can be assigned through the validation process. These help to understand a method’s specificity. Depending on the nature of the method and its applications, criteria for what constitutes “acceptable” performance can be discussed. Three major questions arise when a new method is tested and its precision and accuracy are defined: (1) Is the performance acceptable for each desired application? (2) Can the method’s performance be improved? (3) What will be the performance of the method if it is used in a wide variety of laboratories?
EPA and Association of Analytical Communities (AOAC International) Methods Validation Approaches
The EPA uses a number of different procedures for microbiological analysis of water and other environmental samples. Internally, EPA has established a methods approval streamlining process, called the 1600 Series, to develop, evaluate, and standardize methods designed to detect microorganisms in water and other environmental media. Through this process, Series 1600 methods have been developed and published for Giardia, Cryptosporidium, coliphages, enterococci, Aeromonas, and other microorganisms. In addition, through recent legislation, EPA has attempted to identify, approve, and accept microbiological methods developed and evaluated by entities other than the agency (EPA, 2002). In a recent publication, EPA (2002) approved updated versions of analytical methods developed by the ASTM, those included in Standard Methods, the U.S. Geological Survey, and the U.S. Department of Energy for use in various CWA and SDWA compliance monitoring programs. These latest approvals included multiple editions of the same method, which the agency believes will benefit the regulatory and regulated communities by increasing method selection flexibility and by allowing the continued use of time-tested procedures.
In 1996, EPA proposed a regulation to streamline the program for approving laboratory test procedures and quality control measures that are used to gather data and monitor compliance under the CWA and the SDWA. This effort was geared toward reducing the regulatory burden imposed on industries and municipalities, and the technology development and laboratory services communities. It is also proposed to lower the barriers to innovative technology. A draft Guide to Method Flexibility and Approval of EPA Water Methods was released in 1996 as a result of this effort. However, this report was never finalized; it has not been updated since its original release, and there are no plans to do so in the near future (Lisa Almodovar, EPA, personal communication, 2003). While this proposed program placed the burden of the cost and time for implementation of a new measurement method on industry and laboratories, it provided little incentive for development of new methods or modification of current methods for application to a new matrix. It also focused on compliance monitoring and thus (for example) did not provide incentives for testing methods to address microorganisms on the 1998 Drinking Water Contaminant Candidate List (EPA, 1998; see also Chapter 6).
The development and standardization of new chemical measurement methods has had AOAC International behind it for more than 100 years, with significant funding from several governmental agencies, though primarily the U.S. Department of Agriculture and the U.S. Food and Drug Administration. There are three main programs within AOAC (see Table 5-2 and www.aoac.org for further information):
-
The Official Methods of AnalysisSM program of AOAC provides multilaboratory validation for nonproprietary and commercial proprietary methods where the highest degree of confidence in performance is required to generate credible, defensible, and reproducible results.
-
The Peer-VerifiedSM methods program provides independent laboratory validation for nonproprietary methods where rapid validation and some degree of confidence in performance are needed.
-
The Performance TestedSM methods program provides similar independent laboratory validation of performance claims but for commercial proprietary methods where rapid validation and some degree of confidence are also needed.
Common elements of these three AOAC programs include the following:
-
typical validation time of 12 months minimum;
-
minimum of 8-10 independent labs;
-
in-house method validation data review;
-
publication of methods in the Official Methods of Analysis of AOAC INTERNATIONAL and the Journal of AOAC INTERNATIONAL;
-
citing of methods in U.S. Code of Federal Regulations; and
-
permission of proprietary methods or rapid methods.
The committee concludes that the AOAC’s Peer-Verified Methods program may provide a good approach for the assessment and validation of new microbiological methods in an efficient and defensible manner since its intent is to categorize a group of tested methods that have not yet been subjected to a full collaborative study.
The codification of a method sufficient to reach widespread acceptance and use requires acceptable reproducibility among laboratories and acceptable (and known) levels of false positives and false negatives. In addition, it has long been known that the analysis of truly replicate samples (e.g., aliquots from a well mixed homogenous suspension of microorganisms) should result in observations that are distributed in accordance with Poisson statistics (Armitage and Spicer, 1956; Ziegler and Halvorson, 1935). It is likely that any new method that achieves wide acceptance will need to go through one of the official standardization processes. In this regard, the testing (number of labs, samples, etc.) and evaluation process is likely to be similar (or even more rigorous) than that undertaken to include coliform presence-absence tests in an earlier edition of Standard Methods (Clark and el-Shaarawi, 1993; Pipes et al., 1986).
Recent and ongoing major developments in new methods in microbiology with applications for public health-related water quality have necessitated a new approach for their rapid assessment, standardization, and validation. For example, a recent review on pollution of coastal waters by Griffin and colleagues (2003) found more than nine articles published in the last few years that all used PCR-
based methods for rapid detection of viruses with a variety of specificity and sensitivity but little evidence of validation. Water utilities are purchasing PCR or quantitative PCR machines with funds associated with the new “water security” measures, yet the methods and applications for their effective use are not easily obtainable. It is clear that a major effort is needed in the area of methods for the examination of microbial water quality and health. To move new methods into those deemed conventional, a process is required that not only allows for standardization but also for implementation of the methods so that widespread acceptance and use can result. In addition, there is a need to develop standard approaches for interpreting results, particularly results coming from non-culture-based methods.
While the committee concludes that an AOAC Peer-Verified approach, or its equivalent, may be the best way forward, a major program on methods development must be established with water research laboratories in academic institutions in collaboration with industry research and government research laboratories. The committee recommends that such a program for the development and validation of microbiological analysis of water contain several elements:
-
EPA should strengthen its current role and appropriate interactions and partnerships with standard-setting organizations, including ASTM, AOAC, and ISO that are largely individually driven, to facilitate microbial methods development and help focus on new and innovative methods. In addition, regular and ongoing involvement of professional organizations such as the American Society for Microbiology can bring credible independent third party input.
-
A nationwide database should be created that compiles and serves as a clearinghouse for all microbiological methods that have been utilized and published for studying water quality. Research methods, particularly those that have great potential for evolving to conventional methods, will need to be documented. The funding of methods development has been limited, especially for new and emerging methods and innovative indicators. Rather, the development of new and improved methods has been substantially funded (largely by EPA and the American Water Works Association Research Foundation; AWWARF) for only a few pathogens, specifically those targeted for regulations in drinking water such as Cryptosporidium and Giardia. Greater efforts are required to support methods development for new and emerging microbial detection technologies, for more pathogens, and for new and improved candidate indicators of waterborne pathogens. Approaches for the development and maintenance of an on-line database of new microbiological methods for the analysis of water should be investigated. Guidance on the appropriate data needed for methods studies should be included in this database, and a method for iterative development of consensus methods on-line should be provided.
-
A specific program on promising research methodologies should be sup-
-
ported by EPA. These methodologies need not be microorganism specific, but should be application specific, focusing on the desirable attributes of the method.
CHALLENGES AND PITFALLS OF NEW TECHNOLOGY
A variety of challenges and potential pitfalls will be associated with developing and using improved and rapid methods for microbiological water quality compared with continued use and reliance on time-tested, widely accepted, traditional microbiological methods. These are discussed below.
Scientific Principles in Identifying a Culture
Many new methods employing current and innovative technologies may be more sensitive than the classical methods. However, when there are discrepancies between new (e.g., PCR) and classical methods in identifying a culture (e.g., Salmonella in food and water), the decision as to which method is correct will likely center around determining the identity of the unknown culture. In such situations, the issue of phenotypic expression of cells versus genotypic composition of cells becomes very crucial. Another important comparison issue revolves around the statistical treatment of information. For classical methods, a few attributes will be used to pinpoint the identity of an unknown culture; however, in modern diagnostic kits that utilize many method attributes, whether an unknown culture is identified as Salmonella 90 or 85 percent accurately makes the identification rather ambiguous.
As noted previously, an important question concerns the viability and infectivity of the cell being monitored. Some molecular techniques such as PCR can amplify the DNA of a dead cell and give a positive response concerning the presence of a pathogen in a sample that might be perfectly safe. The different scientific principles involved in determining the identity of a culture deserves more research and debate.
Physical and Separation Issues
Many new biological measurement methods are becoming increasingly miniaturized, computerized, and automated. As the technology for making biological measurements continues to scale down to micro-scale dimensions, the corresponding sample sizes required for such analyses similarly decrease (e.g., submicroliter samples are easily assessed using many of today’s technologies) and the introduction of a target pathogen or indicator organism from a water sample to interact with these extremely small entities (e.g., a million dots of DNA on a microscope slide) becomes a challenge. Furthermore, the use of such small volumes itself poses a serious water sampling problem since the samples presented to the instru-
ment may not contain the species to be measured simply because they are not adequately representative of the bulk sample or because the detection method is not sensitive enough. This sampling problem is of particular concern for pathogens that can pose unacceptable health risks at very low concentrations.
Thus, there is a need to address the sensitivity of miniaturized detection methods and ensure that sample collection, preprocessing or processing, concentration, and purification are given adequate attention. In other words, microorganisms of concern must either be removed from large-volume samples and presented to these miniaturized methods in small aliquots, or larger sample volumes must be passed by the sensor. Currently, this area represents one of the most important technological challenges to the analysis of pathogens and indicators in water and other environmental samples. Separation of target microorganisms from water during sample preparation before application to modern and sophisticated detection systems is an important area for further research. Specifically, elimination, reduction, and destruction of inhibitors, debris, food particles, lipids, proteins, organic and inorganic particles, cellular matter, and so forth, in samples are all important issues to be resolved.
Cost and Technology Transfer
Many modern diagnostic and detection systems utilize sophisticated instrumentation that may be excessively costly for most potential users. In fact, the average cost of an automated instrument can easily reach or surpass $30,000 and perhaps even $250,000 for a mass spectrometer (e.g., Fourier transform infrared spectrometer). Of course, if one performs a large number of tests regularly, the average cost per test will be low, but in many instances, smaller laboratories may find that the volume of tests does not justify the cost of the instrument. For example, a laboratory that routinely conducts less than 100 Salmonella tests per week has little or no need for a sophisticated, automated, and very expensive instrument that can perform thousands of tests per week. The committee recognizes the lack of technical, infrastructure, and financial resources required to implement water monitoring in many parts of the United States and recommends that efforts be made to support the development of inexpensive and rapid fieldable methods for testing microbial water quality.
Finally, while many detection technologies exist that are applicable to the detection of waterborne pathogens and indicator organisms, they are primarily laboratory based. The need to develop rapid fieldable methods will require the concurrent development of reagents, methods, and the attendant portable instruments that can survive repeated transport and use in the less stringently controlled environments of the field.
Unrealistic Expectations and Resistance to Change
New users of an automated measurement device often expect the system to operate perfectly and perform all their necessary tests immediately. Usually, such expectations are too high. Once it is discovered that a particular system does not satisfy all immediate measurement needs, some users will either discard the system totally or develop a negative impression of the system. Users have to understand that an instrument is designed and marketed after extensive testing for specific applications and that even a slight deviation from the specified protocol (e.g., putting acidic water into a sensor not designed to handle a low-pH sample) may result in unsatisfactory performance. No system is 100 percent perfect all the time. At present, there is no sensor that can be placed into a water sample and left alone to make an autonomous measurement without some level of attention.
There is also an intrinsic resistance to change that pervades virtually every analytical community where certain well-established methods have been employed successfully for long periods. This innate conservatism is well founded in some cases where new methods have not been validated. A specific application can sometimes lead to errors or compromise existing long-term data sets. In such cases, it is important that new methods be tested side by side with well-established methods so that the user can acquire a degree of comfort with the new method. The ideal situation is to design “foolproof” systems so that no human error can interfere with the operation of the system from the point of sample application to the end results. Although the microbiological community is moving ever closer to that reality, it is not yet achievable.
LOOK TO THE FUTURE
Today’s measurement techniques are aimed at detecting viable organisms or specific components present in the organism of interest and correlating their presence to human health risk assessment.
Sensors and biosensors are beginning to play a role in several application areas including clinical medicine, environmental science, and process control fields. Analyzers are designed to integrate the steps of sampling, preprocessing, and measurement into a single, functional device. In some cases, sampling is determined by sensor placement; no preprocessing is required due to the sensor’s specificity; signal transduction-detection is an integral function; and sensors offer the potential for real-time monitoring capability because they measure continuously. Further advances in sensors, including making them sufficiently robust for field deployment, will enable them to address some of the measurements discussed throughout this chapter.
Another area that will have an important impact on microbial analysis is molecular recognition. The use of combinatorial methods such as phage display (Sidhu, 2000), aptamers (O’Sullivan, 2002), and combinatorial chemistry has
expanded the ability to rapidly and efficiently generate and screen new molecular entities that may be helpful in producing new recognition elements that can be used as labels, purification reagents, and sensors.
A technology area that will enable significant reductions in sample preparation and separation times is in the field of microfluidics and microelectromechanical systems (MEMS). Complete “lab-on-a-chip” devices are being created out of inexpensive materials such as glass or plastic. These chips contain fully integrated analytical systems with the ability to concentrate, separate, and detect a multitude of analytes including nucleic acids, proteins, and small molecules. Because the overall device sizes are small compared to most benchtop analytical systems, they can perform analysis in second-to-minute time frames. Further advances in chip design and detection schemes should facilitate more complex and sensitive analysis.
One of the most exciting fields of current research in science and technology is the area of nanotechnology. In this area, defined as systems with features on the nanometer scale, functional devices and materials are being developed at an increasing rate. While the material costs associated with the technology may be high, the number of devices one can prepare from a small amount of material is enormous. For example, a gram of nanoparticles contains trillions of individual particles, each of which can, in principle, serve a particular function.
With the advent of nanotechnology, and even microtechnology, materials costs will therefore actually decrease. The ability to pack functions, such as communications hardware, on-board processing, and signal transduction, into ever-smaller devices suggests that in the not-too-distant future it should be possible to create sensors with a high degree of measurement capability in an extremely small device.
One of the more recent trends in sensing systems is array technology (see also Appendix C). In these systems, tens to thousands of sensor elements can be placed on a single substrate with overall dimensions of several square millimeters. The burgeoning area of DNA microarrays for genomics is driving advances in this area. Developments in protein and carbohydrate arrays will further advance the applicability of arrays to microbial analysis. Nanotechnology will cause feature sizes to shrink even further. The ability to place so many sensors on a single device raises the prospect of what has been referred to as a “universal sensor”—a system able to detect virtually anything of interest. Such systems can be built on chips in which a sensor is present for every analyte of potential interest. Alternatively, such arrays may be able to measure patterns of response in which signatures of various analytes signify the presence of various water quality conditions, organisms, or toxins. In this approach, pattern recognition algorithms, combined with prior training, could be used to assess water quality and identify potential hazards. One of the advantages of such a system is its ability to be anticipatory, such that new or difficult-to-culture pathogens could be detected by presenting a signature that is similar to known pathogens.
As described previously and elaborated further in the Appendix C, mass spectral techniques for performing microbial analysis using entire mass spectral patterns is in its infancy and should also have an impact, with the limitation that it is unlikely to be an inexpensive, portable field analytical tool.
With the prospect for such an enormous amount of data to be collected from the many sensors disposed on arrays, the large numbers of sensor arrays deployed for water monitoring, and the continuous data streams coming from these sensor networks, attention must be paid to data analysis, intelligent decision making, and archiving.
Ongoing research in the micro- and nanotechnology field, combined with efforts in array sensing and intelligent processing, should provide the tools for creating inexpensive, ubiquitous, universal sensing and detection systems beginning now and continuing over the next several decades.
While many of the new and innovative molecular methods discussed in this chapter (and Appendix C) enhance the opportunity for direct measurement of pathogens, more effective use of direct pathogen measurement will require establishment of the correlation between pathogen concentration and health risk. There are presently no standards on which to base health risk decisions for most pathogens. Current epidemiologic studies (as reviewed elsewhere in this report), on which recreational water exposure standards are based, have been conducted almost exclusively for indicator bacteria such as fecal coliforms and enterococci.
Even for presently used indicator bacteria, the relationship to health risk will have to be reestablished for the new molecular-based methods. Existing epidemiology studies have all been based on quantifying exposure using culture-based methods, which measure some aspect of metabolic activity. Some of the new indicator and pathogen methods quantify the presence of cellular structure, but many do not assess the ability to grow or to infect. As such, they have the potential to overestimate health risk relative to present standards.
Consistent with its previous related recommendations, the committee recommends that epidemiologic studies should be designed and performed to both establish the correlation among indicator and pathogen concentrations and health risk, and reestablish the health risks associated with existing and new pathogen indicators for new (non-culture-based) detection methods.
SUMMARY: CONCLUSIONS AND RECOMMENDATIONS
Recent, emerging, and forecasted advances in microbiology, molecular biology, and analytical chemistry make it timely to reassess the long-standing paradigm of relying primarily or exclusively on traditional microbial (primarily bacterial) indicators for waterborne pathogens to support public health decision making regarding the microbiological quality of water. Although classic microbiological culture methods for detection of indicator microorganisms and pathogens have proved effective over many decades, they suffer from a number of
limitations that are discussed throughout this report. The advent of increasingly sophisticated and powerful molecular biology techniques provides new opportunities to improve upon present indicators and pathogens by both culture and non-culture methods. What is needed is a phased monitoring approach that makes use of a flexible tool box, in which a variety of indicator methods and approaches are available for measuring a given indicator or pathogen for different applications that considers spatial and temporal scale (granularity) issues. The need for such a phased monitoring approach and examples of its implementation are discussed in detail in Chapter 6.
It is vital that all four components of the process of performing a measurement (i.e., collection, sample processing or preprocessing, measurement, and data processing; not just the measurement itself) be considered in order to make an accurate analysis of microbial water quality. The collection of representative samples requires careful consideration of the objectives or purpose of sampling in the context of the need to obtain a reliable estimate of microbial exposure in a timely fashion. Furthermore, the widespread use of logarithmic transformations and measure of central tendency and dispersion of log-transformed data to estimate exposures and health risks needs to be reconsidered in water microbiology, epidemiology, and health risk assessment.
At present, most water quality measurement methods are single-parameter based. Ongoing research in the micro- and nanotechnology field, combined with efforts in array sensing and intelligent processing, should provide the tools for creating inexpensive, ubiquitous universal sensing and detection systems beginning now and over the next several decades. This development is essential because the committee recognizes the lack of technical, infrastructure, and financial resources required to implement advanced water quality monitoring methods in many parts of the United States. The microbiological community needs to develop and implement multiparameter approaches in which many technologies and methods are integrated to provide the best possible information. Similarly, the water monitoring community needs to be aware of new developments in these areas that can be brought to bear on microbiological water quality monitoring.
Although evolving detection methods will be increasingly able to rapidly detect specific pathogens, the use of well-characterized (conventional) indicator approaches will continue to be necessary because our understanding of existing and emerging pathogens will never be complete. Regardless, more effective use of direct pathogen measurement discussed in this chapter will require establishment of the relationship between pathogen concentration and health risk (see also Chapters 2 and 4). Similarly, the relationship to health risk will have to be reestablished for presently used indicator bacteria and new (non-culture-based) methods.
The funding of methods development has been relatively poor to date for many pathogens, for new and emerging methods, and for new and innovative indicators. Investigations into only a few pathogens, specifically those targeted
for regulations in drinking water such as Cryptosporidium and Giardia, have been substantially funded (largely by EPA and AWWARF) for the development of new and improved methods. Greater and more consistent efforts should be made to support methods development for new and emerging microbial detection technologies, for many more pathogens, and for new and improved candidate indicators of waterborne pathogens.
Newer methods involving immunofluorescence techniques and nucleic acid analysis are proving their value, and novel microtechnologies are evolving rapidly, spurred in part by recent concerns about bioterrorism. Problems associated with sample concentration, purification, and efficient (quantitative) recovery remain and will require significant effort to be resolved. One technology area that will enable significant reductions in sample preparation and separations time is the field of microfluidics and MEMS. Thus, the introduction of molecular techniques for nucleic acid analysis is viewed by the committee as a growth opportunity for waterborne pathogen detection.
With the prospect for such an enormous amount of data to be collected from the many sensors disposed on arrays, the potentially large numbers of sensor arrays deployed for water monitoring, and the continuous data streams coming from these sensor networks, greater attention must be paid to the fields of data analysis, intelligent decision making, and archiving. There is need for a database that compiles and serves as a clearinghouse for all microbiological methods that have been utilized and published for studying water quality. Research methods, in particular those that have great potential for evolving into conventional methods, will have to be documented.
Recent developments in molecular and microbiology methods and their application to public health-related water microbiology have necessitated a new approach for rapid assessment, standardization, and validation of such methods. It is clear that a major effort is needed for accessible methods to examine microbial water quality for health decisions. To move new methods into the main-stream, a process is required that not only allows for standardization and validation but also facilitates widespread acceptance and implementation. In this regard, the committee concludes that the AOAC Peer-Verified approach or its equivalent may be the best way forward. However, a major program on methods development will need to be established with water research laboratories in academic institutions in collaboration with industry research and government research laboratories.
Based on these conclusions, the committee makes the following recommendations:
-
A specific program on promising research methodologies for waterborne microorganisms of public health concern should be supported by EPA and other organizations concerned with microbial water quality. Such methodologies need
-
not be microorganism specific, but should be application specific, focusing on the desirable attributes of the method.
-
Ongoing research should be supported and expanded to develop and validate rapid, sensitive, and robust methods for detection and measurement of all classes of waterborne pathogens and their indicators. Such expanded research should go beyond pathogenic bacteria and indicators, to include improved methods for the detection of pathogenic viruses and protozoa.
-
Additional research is needed to develop improved methods for rapid sample concentration and effective, reproducible microbial recovery. Specifically, elimination, reduction, and destruction of inhibitors, debris, food particles, lipids, proteins, organic and inorganic particles, cellular matter, and so forth, in samples are important issues.
-
Research should be funded to develop approaches to the detection of infectious or viable microbes by nucleic acid detection methods, including the use of ligand-binding steps in microbial recovery from samples to select for intact and infectious microbes.
-
The adoption of new molecular techniques should be accelerated for waterborne pathogen detection. New methods undergoing validation should be tested using whole microorganisms, rather than just extracted DNA or RNA targets, to perform tests for sensitivity and linearity.
-
Focused efforts should be made to support the development of inexpensive and rapid fieldable methods for testing microbial water quality. This will require the concurrent development of reagents, methods, and the attendant portable instruments that can survive repeated transport and use in the field.
-
There is a need to address the sensitivity of miniaturized detection methods and ensure that sample collection, preprocessing or processing, concentration, and purification are given adequate attention to achieve representativeness and have the ability to detect microbial concentrations posing unacceptable health risks. This represents one of the most important technological challenges to the analysis of pathogens and indicators in water and other environmental samples and will become more important with the introduction of micro- and nanotechnologies.
-
EPA should reinvigorate its role with standard-setting organizations (including ASTM, AOAC International, and ISO) to facilitate microbial methods development that focuses particularly on new and innovative methods. In addition, regular and ongoing involvement of professional organizations such as the American Society for Microbiology will bring credible, independent, third-party input.
-
EPA should support the design, development, and maintenance of a nationwide database that compiles and serves as a clearinghouse for all microbiological methods that have been utilized and published for studying water quality. Guidance on the appropriate data needed for methods studies should be included
-
in this database. Finally, a means for iterative development of consensus methods on-line should be provided.
-
The committee recommends that epidemiologic studies should be designed and performed to both establish the correlation among indicator and pathogen concentrations and health risk, and reestablish the health risks associated with existing and new pathogen indicators for new (non-culture-based) detection methods.
REFERENCES
APHA (American Public Health Association). 1998. Standard Methods for the Examination of Water and Wastewater, 20th Edition. Washington, D.C.
Armitage, P., and C.C. Spicer. 1956. The detection of variation in host susceptibility in dilution counting experiments. Journal of Hygiene 54: 401-414.
Barnhard, A.E., and K.G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Applied and Environmental Microbiology 66(4): 1587-1594.
Blackmer, F., K.A. Reynolds, C.P. Gerba, and I.L. Pepper. 2000. Use of integrated cell culture-PCR to evaluate the effectiveness of poliovirus inactivation by chlorine. Applied and Environmental Microbiology 66(5): 2267-2268.
Bogosian, G., and E.V. Bourneuf. 2001. A matter of bacteria life and death. EMBO Reports 2(9): 770-774.
Bogosian, G., P.J. Morris, and J.P. O’Neil. 1998. A mixed culture recovery method indicates that enteric bacteria do not enter the viable but nonculturable state. Applied and Environmental Microbiology 64(5): 1736-1742.
Breaker, R.R. 2002. Engineered allosteric ribozymes as biosensor components. Current Opinion in Biotechnology 13(1): 31-39.
Call, J.L., M. Arrowood, L.T. Xie, K. Hancock, and V.C.W. Tsang. 2001. Immunoassay for viable Cryptosporidium parvum oocysts in turbid environmental water samples. Journal of Parasitology 87: 203-210.
Chandler, D.P., J. Brown, D.R. Call, J.W. Grate, D.A. Holman, L. Olson, M.S. Stottlmyre, and C.J. Bruckner-Lea. 2001. Continuous, automated immunomagnetic separation and microarray detection of E. coli O157:H7 from poultry carcass rinse. International Journal of Food Microbiology 70: 143-154.
Clark, J.A., and A.H. el-Shaarawi. 1993. Evaluation of commercial presence-absence test kits for detection of total coliforms, Escherichia coli, and other indicator bacteria. Applied and Environmental Microbiology 59: 380-388.
Collier, J. L., and L. Campbell. 1999. Flow cytometry in molecular aquatic ecology. Hydrobiologia 401: 33-53.
Colwell, R.R., and D.J. Grimes, eds. 2000. Nonculturable Microorganisms in the Environment. Washington, D.C.: American Society for Microbiology Press.
Colwell, R.R., P. Brayton, A. Huq, B. Tall, P. Harrington, and M. Levine. 1996. Viable but nonculturable Vibrio cholerae O1 revert to a culturable state in the human intestine. World Journal of Microbiology and Biotechnology 12: 28-31.
Cook, N. 2003. The use of NASBA for the detection of microbial pathogens in food and environmental samples. Journal of Microbiological Methods 53(2): 165-174.
Deininger, R.A., and J. Lee. 2001. Rapid determination of bacteria in drinking water using an ATP assay. Field Analytical Chemistry and Technology 5(4): 185-189.
DiGiovanni, G.D., F.H. Hashemi, N.J. Shaw, F.A. Abrams, M.W. LeChevallier, and M. Abbaszadegan. 1999. Detection of infectious Cryptosporidium parvum oocysts in surface and filter backwash water samples by immunomagnetic separation and integrated cell culture-PCR. Applied and Environmental Microbiology 65(8): 3427-3432.
Dombeck, P.E., L.K. Johnson, S.T. Zimmerly, and M.J. Sadowsky. 2000. Use of repetitive DNA sequences and the PCR to differentiate Escherichia coli isolates from human and animal sources. Applied and Environmental Microbiology 66(6): 2572-2577.
Donaldson, K.A., D.W. Griffith, and J.H. Paul. 2002. Detection, quantitation, and identification of enteroviruses from surface waters and sponge tissue from the Florida Keys using real-time RT-PCR. Water Research 36(10): 2505-2514.
Dore, W.J., K. Henshilwood, and D.N. Lees. 2000. Evaluation of F-specific RNA bacteriophage as a candidate human enteric virus indicator for bivalve molluscan shellfish. Applied and Environmental Microbiology 66(4): 1280-1285.
Dore, W.J., M. Mackie, and D.N. Lees. 2003. Levels of male-specific RNA bacteriophage and Escherichia coli in molluscan bivalve shellfish from commercial harvesting areas. Letters in Applied Microbiology 36(2): 92-96.
Edwards, C. 2000. Problems posed by natural environments for monitoring microorganisms. Molecular Biotechnology 15(3): 211-223.
EPA (U.S. Environmental Protection Agency). 1996. Guide to Method Flexibility and Approval of EPA Water Methods (draft). Washington, D.C.: Office of Water.
EPA. 1998. Announcement of the Drinking Water Contaminant Candidate List. Notice. Federal Register 64(40): 10274-10287.
EPA. 2000. National Primary Drinking Water Regulations: Ground Water Rule; Proposed Rule. Federal Register 65(91): 30193-30274.
EPA. 2002. Guidelines Establishing Test Procedures for the Analysis of Pollutants Under the Clean Water Act; National Primary Drinking Water Regulations; and National Secondary Drinking Water Regulations; Methods Update. Federal Register 67(205): 65219-65253.
Fleischer, J.M., F. Jones, D. Kay, R. Stanwell-Smith, M. Wyer, and R. Morano. 1993. Water and nonwater related risk factors for gastroenteritis among bathers exposed to sewage contaminated marine waters. International Journal of Epidemiology 22(4): 698-708.
Feng, S., and P.T.K. Woo. 2001. Cell membrane glycoconjugates on virulent and avirulent strains of the haemoflagellate Cryptobia salmositica (Kinetoplastida). Journal of Fish Diseases 24(1): 23-32.
Fung, D.Y.C. 2002. Rapid methods and automation in microbiology. Comprehensive Reviews in Food Science and Food Safety 1: 1-20.
Gehring, A.G., C.G. Crawford, R.S. Mazenko, L.J. Vanhouten, and J.D. Brewster. 1996. Enzyme-linked immunomagnetic electrochemical detection of Salmonella typhimurium. Journal of Immunological Methods 195(1-2): 15-25.
Griffin, D.W., K.A. Donaldson, J.H. Paul, and J.B. Rose. 2003. Pathogenic human viruses in coastal waters. Clinical Microbiology Reviews 16(1): 129-143.
Hald, B., K. Knudsen, P. Lind, and M. Madsen. 2001. Study of the infectivity of saline-stored Campylobacter jejuni for day-old chicks. Applied and Environmental Microbiology 67(5): 2388-2392.
Huffman, D.E., T.R. Slifko, M.J. Arrowood, and J.B. Rose. 2000. Inactivation of bacteria, virus, and Cryptosporidium by a point-of-use device using pulsed broad spectrum white light. Water Resources 34(9): 2491-2498.
Hurst, A. 1977. Bacterial injury: A review. Canadian Journal of Microbiology 23: 936-944.
Hurst, C.J., G.R. Knudsen, M.J. McInerney, L.D. Stetzenbach, and M.V. Walter, eds. 2002. Manual of Environmental Microbiology. Washington, D.C.: American Society for Microbiology Press.
Ishida, Y., A.J. Madonna, J.C. Rees, M.A. Meetani, and K.J. Voorhees. 2002. Rapid analysis of intact phospholipids from whole bacterial cells by matrix-assisted laser desorption/ionization mass spectrometry combined with on-probe sample pretreatment. Rapid Communications in Mass Spectrometry 16(19): 1877-1882.
Jones, D.M., E.M. Sutcliffe, and A. Curry. 1991. Recovery of viable but non-culturable Campylobacter jejuni. Journal of General Microbiology 137(Part 10): 2477-2482.
Kay, D., J.M. Fleischer, R.I. Salomon, F. Jones, M.D. Wyer, A.F. Godfree, Z. Zelenauch-Jacquotte, and R. Shore. 1994. Predicting likelihood of gastroenteritis from sea bathing: Results from randomised exposure. Lancet 344: 905-909.
Keer, J.T., and L. Birch. 2003. Molecular methods for the assessment of bacterial viability. Journal of Microbiological Methods 53(2): 175-183.
Kell, D.B., A.S. Kaprelyants, D.H. Weichart, C.R. Harwood, and M.R. Barer. 1998. Viability and activity in readily culturable bacteria: A review and discussion of the practical issues. Antonie Van Leeuwenhoek 73(2): 169-187.
Kilvington, S., and J. Beeching. 1995. Identification and epidemiological typing of Naegleria fowleri with DNA probes. Applied and Environmental Microbiology 61(6): 2071-2078.
Kreader, C.A. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Applied and Environmental Microbiology 62(3): 1102-1106.
Lammerding, A.M., A.M. Fazil, and G.M. Paoli. 2001. Microbial food safety risk assessment. In Compendium of Methods for the Microbiological Examination of Foods, F.P. Downes and K. Ito, eds. Washington, D.C.: American Public Health Association.
Lee, J., S. Dawson, S. Ward, S.B. Surman, and K.R. Neal. 1997. Bacteriophages are a better indicator of illness rates than bacteria among users of a white water course fed by a lowland river. Water Science and Technology 35(11-12): 165-170.
Lee, Y.M., P.W. Johnson, J.L. Call, M.J. Arrowood, B.W. Furness, S.C. Pichette, K.K. Grady, P. Reeh, L. Mitchell, D. Bergmire-Sweat, W.R. MacKenzie, and V.C.W. Tsang. 2001. Development and application of a quantitative, specific assay for Cryptosporidium parvum oocysts detection in high-turbidity environmental water samples. American Journal of Tropical Medicine and Hygiene 65: 1-9.
Madonna, A.J., K.J. Voorhees, and T.L. Hadfield. 2001. Rapid detection of taxonomically important fatty acid methyl ester and steroid biomarkers using in situ thermal hydrolysis/methylation mass spectrometry (THM-MS): Implications for bioaerosol detection. Journal of Analytical and Applied Pyrolysis 61(1-2): 65-89.
McFeters, G.A., and A.K. Camper. 1983. Enumeration of indicator bacteria exposed to chlorine. Advanced Applied Microbiology 29: 177-193.
McFeters, G.A., M.W. LeChevallier, A. Sing, and J.S. Kippin. 1986a. Health significance and occurrence of injured bacteria in drinking water. Water Science and Technology 18(10): 227-231.
McFeters, G.A., J.S. Kippin, and M.W. LeChevallier. 1986b. Injured coliforms in drinking water. Applied and Environmental Microbiology 51: 1-5.
Medema, G.J., F.M. Schets, A.W. van de Giessen, and A.H. Havelaar. 1992. Lack of colonization of 1 day old chicks by viable, non-culturable Campylobacter jejuni. Journal of Applied Bacteriology 72(6): 512-516.
Mitchell, B.A., J.A. Milbury, A.M. Brookins, and B.J. Jackson. 1994. Use of immunomagnetic capture on beads to recover Listeria from environmental samples. Journal of Food Protection 57(8): 743-745.
Murray, P.R., E.J. Baron, M.A. Pfaller, F.C. Tenover, and R.H. Yolken, eds. 1999. Manual of Clinical Microbiology, 7th Edition. Washington, D.C.: American Society for Microbiology Press.
Nuanualsuwan, S., and D.O. Cliver. 2002. Pretreatment to avoid positive RT-PCR results with inactivated viruses. Journal of Virological Methods 104(2): 217-225.
O’Sullivan, C.K. 2002. Aptasensors: The future of biosensing. Analytical and Bioanalytical Chemistry 372(1): 44-48.
Paoli, G. 2002. Microbial risk assessment: Lessons learned and future directions. Presented at 1st International Conference on Microbiological Risk Assessment: Foodborne Hazards. College Park, Maryland: University of Maryland.
Pipes, W.O., H.A. Minnigh, B. Moyer, and M.A. Troy. 1986. Comparison of Clark’s presence-absence test and the membrane filter method for coliform detection in potable water supplies. Applied and Environmental Microbiology 52: 439-443.
Quintero-Betancourt, W., E.M. Peele, and J.B. Rose. 2002. Cryptosporidium parvum and Cyclospora cayetanensis: A review of laboratory methods for detection of these waterborne parasites. Journal of Microbiological Methods 49: 209-224.
Ray, B. 1989. Injured Index and Pathogenic Bacteria: Occurrence and Detection in Foods, Water, and Feeds. Boca Raton, Florida: CRC Press.
Reynolds, K.S., C.P. Gerba, and I.L. Pepper. 1996. Detection of infectious enteroviruses by an integrated cell culture-PCR procedure. Applied and Environmental Microbiology 62(4): 1424-1427.
Reynolds, K.S., C.P. Gerba, and I.L. Pepper. 1997. Rapid PCR-based monitoring of infectious enteroviruses in drinking water. Water Science and Technology 35(11-12): 423-427.
Rose, J.B., D.E. Huffman, and A. Gennaccaro. 2002. Risk and control of waterborne cryptosporidiosis. FEMS Microbiology Reviews 26: 113-123.
Sidhu, S.S. 2000. Phage display in pharmaceutical biotechnology. Current Opinion in Biotechnology 11(6): 610-616.
Slifko, T.R., D.E. Huffman, D. Bertrand, J.H. Owens, W. Jakubowski, C.N. Haas, and J.B. Rose. 2002. Comparison of animal infectivity and cell culture systems for evaluation of Cryptosporidium parvum oocycts. Experimental Parasitology 101: 97-106.
Smith, R.J., A.T. Newton, C.R. Harwood, and M.R. Barer. 2002. Active but nonculturable cells of Salmonella enterica serovar Typhimurium do not infect or colonize mice. Microbiology 148(Part 9): 2717-2726.
Sobsey, M.D., K.J. Schwab, and T.R. Handzel. 1990. A simple membrane filter method to concentrate and enumerate male-specific RNA coliphages. Journal of the American Water Works Association 82(9): 52-59.
Sobsey, M.D., D.A. Battigelli, G.A. Shin, and S. Newland. 1998. RT-PCR amplification detects inactivated viruses in water and wastewater. Water Science and Technology 38(12): 91-94.
Sobsey, M.D. 1999. Methods to identify and detect microbial contaminants in drinking water. Pp. 173-205 in Identifying Future Drinking Water Contaminants. Washington, D.C.: National Academy Press.
Sobsey, M.D. 2001. Microbial detection: Implications for exposure, health effects, and control. Pp 89-113 in Microbial Pathogens and Disinfection By-Products in Drinking Water: Health Effects and Management of Risks, G.F. Craun, F.S. Hauchman, and D.E. Robinson, eds. Washington, D.C.: ILSI Press.
Veal, D.A., D. Deere, B. Ferrari, J. Piper, and P.V. Attfield. 2000. Fluorescence staining and flow cytometry for monitoring microbial cells. Journal of Immunological Methods 243(1-2): 191-210.
Zhen, B., Y.J. Song, Z.B. Guo, J. Wang, M.L. Zhang, S.Y. Yu, and R.F. Yang. 2002. In vitro selection and affinity function of the aptamers to Bacillus anthracis spores by SELEX. Acta Biochimica et Biophysica Sinica 34(5): 635-642.
Ziegler, N.R., and H.G. Halvorson. 1935. Application of statistics to problems in bacteriology. IV. Experimental comparison of the dilution method, the plate count, and the direct count for the determination of bacterial populations. Journal of Bacteriology 29: 609-634.















































