5
Emerging Technical Tools
OVERVIEW
A trend that has long been recognized is that people who have had influenza may have less severe symptoms when subsequently infected with immunologically distinct viruses. Immunization with the virus of one influenza A subtype has been shown to reduce morbidity and mortality in animals infected with virus of a different subtype, a phenomenon known as heterosubtypic immunity. In the first contribution to this chapter, Suzanne Epstein describes animal studies on the various means of inducing heterosubtypic immunity and explores the possibility of taking advantage of conserved features among influenza viruses to reduce mortality in a pandemic until a matched vaccine became widely available. Routine immunization could potentially be used to induce heterosubtypic immunity in advance of a pandemic, and the vaccine could also be offered early in a pandemic to those who had not received it.
An even more ambitious strategy is presented in the next contribution, which describes the engineering of influenza A-resistant chickens that combines (1) RNA interference, (2) genes that block the expression of incoming viral genomes, and (3) RNA decoys, short sequences that mimic the binding sites of RNA proteins and thereby act as competitive inhibitors for transcription. Although researchers pursing this strategy recognize the many logistical and scientific roadblocks in their path, they nonetheless envision the elimination of a major pandemic threat through global repopulation with influenza-resistant transgenic chickens.
Rapid detection techniques are critically needed for a quick diagnosis
of the pathogen. The faster the pathogen is detected, the faster the outbreak can be controlled. The chapter continues with a description of several novel approaches for rapid early detection, including the most promising assay, real-time fluorescent polymerase chain reaction, as well as some other techniques: antigen capture/enzyme-linked immunosorbent assay, mass spectrometry, and restriction fragment length polymorphisms. The development of these techniques for detection will enable a quick diagnosis of the agent and faster development of vaccines.
The chapter concludes with mathematical modeling of pandemic preparedness plans, showing the consequences on health economic outcomes of possible intervention strategies. This modeling helps to determine the costs and benefits of different strategies and gauges the public health benefits of optimized preparedness.
CONTROL OF INFLUENZA VIRUS INFECTION BY IMMUNITY TO CONSERVED VIRAL FEATURES
Office of Cellular, Tissue and Gene Therapies, Center for Biologics Evaluation and Research, Food and Drug Administration
Reprinted with permission, from Epstein (2003), published in Expert Review of Anti-infective Therapy, Copyright 2003 Future Drugs
Influenza has circulated among humans for centuries and kills more people than many newly emerging diseases. The present methods for control of influenza are not adequate, especially for dealing with a pandemic. In the face of a rapidly spreading outbreak, a race to isolate the virus and prepare a vaccine would probably not succeed in time to avoid great losses. Thus, additional anti-infection strategies are needed. Broad cross-protection against widely divergent influenza A subtypes is readily achieved in animals by several means of immunization. How does cross-protection work in animals, and can we apply what we have learned about it to induce broad cross-protection in humans?
|
1 |
Suzanne L. Epstein, PhD, Chief, Laboratory of Immunology and Developmental Biology, Division of Cellular and Gene Therapies, HFM-730, Office of Cellular, Tissue and Gene Therapies, Center for Biologics Evaluation and Research, Food and Drug Administration, 301–827–0450, fax: 301–827–0449, e-mail: epsteins@cber.fda.gov. |
|
2 |
SLE acknowledges grant support from the National Vaccine Program. I thank Steven Bauer, Ira Berkower, Mark Tompkins, Zi-Shan Zhao and Chia-Yun Lo for critical review of various versions of the manuscript. |
Background: Influenza Virus, Immunity, and Vaccination
If an influenza pandemic began, emergency efforts to isolate the pandemic virus strain and prepare a vaccine from it would begin, while the pandemic spread. Should we pin our hopes on that race? On antiviral drugs? Or are there vaccines that could be made in advance and offer some degree of protection? Current vaccines focus on variable, strain-specific epitopes of circulating influenza virus strains and new viral strains require new vaccines. Here, a different approach will be considered, vaccination based on shared epitopes as an anti-infective measure that could provide broad protection against even new pandemic strains.
This review will draw together relevant observations from animal studies and from human epidemiology. In the venerable influenza field, some of the older literature is highly relevant to current questions in ways that were not considered at the time, so it must be revisited. After providing background on influenza infection and immunity, the review will focus on broad cross-protection against influenza A subtypes. It will explore the mechanisms of cross-protection in animals, the induction of cross-protection by vaccines of different types and their ability to protect against challenge with potential pandemic subtypes, such as H5N1. Finally, it will consider the possibility of broad immune cross-protection in humans and the public health implications for control of epidemics and pandemics.
Influenza remains a major public health problem. The World Health Organization (WHO) estimates that in a typical year, 10 to 20 percent of the world’s population is infected with influenza, resulting in 3,000,000 to 5,000,000 severe illnesses and 250,000 to 500,000 deaths (World Health Organization, 1999). In the United States, there are tens of thousands of deaths each year and the problem will increase due to the aging of the population and the susceptibility of the elderly.
During pandemics, the losses are even greater. The 1918 influenza pandemic was the most extreme, causing two billion cases, 20 to 40 million deaths worldwide and 500,000 in the United States, and killing with great speed. Young, healthy adults were not spared and approximately 80 percent of the U.S. Army’s World War I deaths were due to influenza (Wright and Webster, 2001). Pandemics in 1957 and 1968 also caused widespread disease and excess deaths. For further historical information, see Kilbourne (1975).
Vaccination is a highly successful strategy for controlling infectious diseases. It is cost-effective and population-wide campaigns are feasible. However, the pathogens against which vaccination has been most successful (e.g., smallpox and polio) have viral types that are few in number and genetically stable. With influenza virus, extensive genetic variation leads to the problem that different dominant viral strains circulate in the human population each year (Figure 5-1). The current vaccine system involves
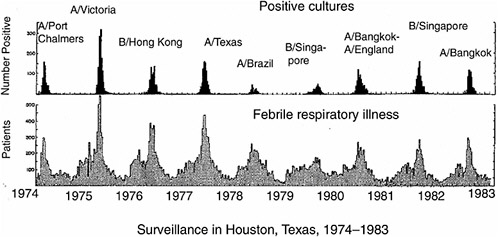
FIGURE 5-1 Periodic outbreaks of influenza in a surveillance study in Texas. Lower panel shows number of persons with acute febrile respiratory illness, upper panel shows number of persons with positive cultures for influenza virus. Strain names are of predominant viruses only. Note that different viral strains dominate in different years. Adapted from Figure 1 in Glezen et al. (1984). Reprinted with permission from Elsevier, Oxford, UK.
worldwide surveillance, predictions of strains likely to circulate during the next season and manufacture of new vaccines. This system is cumbersome, imperfect in effectiveness due to the guesswork involved and rushed in timing. Delays in vaccine derivation and manufacture can lead to shortages, as occurred in 2000 (CDC, 2000). A vaccination strategy that included broad cross-protection in addition to strain-specific protection could have a major public health impact; therefore, the potential of such an approach needs to be thoroughly explored.
There are three major influenza virus types, A, B and C. Infection with influenza C virus is relatively mild clinically (Murphy and Webster, 1990) and will not be discussed here. Influenza A and B viruses are distantly related but not cross-reactive or -protective against each other in animals (de St. Groth and Donnelley, 1950), even during mixed simultaneous infection (Liang et al., 1994). The influenza virus A and B genomes each consist of eight separate RNA segments. Point mutations lead to “antigenic drift” (small, incremental changes). Reassortment of entire segments of the genome is an additional source of antigenic variation and, in the case of influenza A, can lead to “antigenic shift” (sudden, large change) corresponding to a change in subtype.
Influenza virus and its components are shown diagrammatically in Figure 5-2. Hemagglutinin (HA) and neuraminidase (NA) are the components that vary the most. Subtypes of influenza A virus are defined serologically by their HA and NA antigens. The nomenclature for influenza A reflects this, for example, H3N2 refers to HA of subtype 3 and NA of subtype 2. There are 15 HA subtypes and nine NA subtypes (Wright and Webster, 2001). All these subtypes infect aquatic birds, and human pandemic viruses have arisen from avian viruses by reassortment (Webster, 2002). Emergence in humans of subtypes they have not previously encountered can lead to pandemics, for example, the emergence of H1N1 in 1918, H2N2 in 1957 and H3N2 in 1968 (Kilbourne, 1975). Small outbreaks of novel subtypes in humans occur more often than pandemics, for example, H5N1 in Hong Kong in 1997 (Claas et al., 1998), H9N2 in Hong Kong in 1999 (Saito et al., 2001), or an isolated case of H7N7 in The Netherlands in 2003 (van Kolfschooten, 2003).
Immunization with the virus of one influenza A subtype can protect animals against challenge with virus of a different subtype. This cross-protection has long been studied in animal models (Schulman and Kilbourne, 1965). In this review, it will be called heterosubtypic immunity or Het-I, to use the abbreviation of Gerhard (Liang et al., 1994) (it has also been called heterotypic immunity by some authors). This form of immunity does not generally prevent all infection by the heterosubtypic virus but it leads to more rapid viral clearance and to reduction in morbidity and mortality. In this review, the terms ‘protection’ and ‘protective immunity’
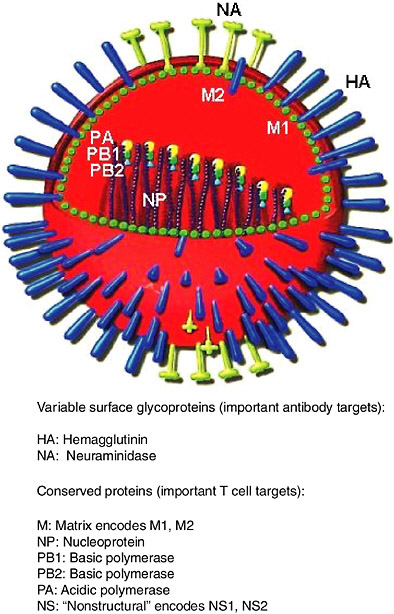
FIGURE 5-2 Diagram of influenza virus and its components. The core containing the RNA genome and replication machinery is surrounded by a matrix and then an envelope. HA, NA and M2 extend through the envelope to the outside. Diagram adapted from (www.snm.ch/public/sante/prevention/prevention-sommarie.htm). Reproduced with permission from Dr. Herve Zender, La Societe Neuchateloise de Medecine (SNM).
will not imply complete prevention of viral infection, but instead, a reduction in viral titers and protection of the life and health of the host.
Figure 5-3 shows diagrammatically the categories of influenza viruses, their relatedness, the terms for describing immunity to various challenges and the resulting protection. Relatedness of viral core and envelope proteins is shown by similarity of color. The corresponding relationships in protein sequence are shown in Table 5-1 for the HA and NA proteins, as well as for nucleoprotein (NP) as an important conserved protein.
A variety of birds and mammals can be infected with influenza A and B viruses, naturally or in the laboratory (Kilbourne, 1987). Serological reagents are often produced in ferrets. Influenza viruses can be adapted to
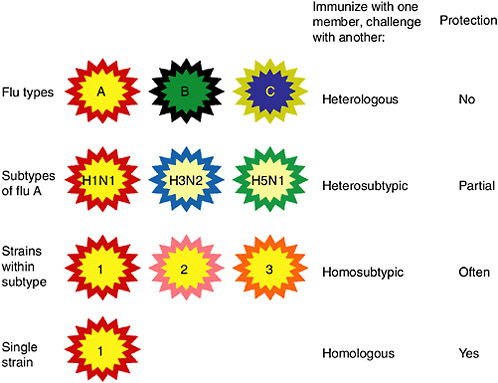
FIGURE 5-3 Categories of influenza viruses and immunity they induce. Colors indicate similarity. For example, influenza virus types A, B and C differ for both internal proteins and the HA and NA external glycoproteins. Within the influenza A type, subtypes have major differences in HA and NA but only subtle differences in internal proteins. Within an influenza A subtype, for example, H1N1, the HA and NA differ in more subtle ways shown by the more similar colors. Note that the term ‘heterologous’ immunity is also used to refer to immunity induced by one virus and reactive with an unrelated virus (Selin et al., 1994) but the term will not be used that way in this review.
TABLE 5-1 Influenza Viruses: Nomenclature and Relatedness
mice and in them cause disease with many of the characteristics of human influenza: upper respiratory infection, tracheobronchitis and pneumonia (Yetter et al., 1980; Renegar, 1992). They provide an affordable animal model with a short generation time and many reagents defining surface markers on important cellular populations. In addition, there are numerous recombinant and congenic strains, and more recently transgenic and knockout strains of immunological significance. Thus, much work on immunity to influenza virus infection has been performed in mice. Results in an animal model do not predict in every particular what will happen in humans, but they provide a valuable information base that can help design future studies in humans and novel approaches to vaccine development.
Immunity to Influenza Virus Infection
How to Analyze Mechanisms of Immunity in Animal Models
The complexity and redundancy of the immune system is good for defense against pathogens but hard on those trying to interpret experiments. Any response that is measured was likely accompanied by other concurrent responses that were not. Thus, correlation of a response with protection does not prove that it mediates the protection. Passive transfer of antibody or T-cells helps by showing that an effector is capable of mediating an outcome, but does not mean that it always does. The adoptive
transfer may use unnatural doses and the transferred components may not localize normally.
Mice with a targeted gene disruption (‘knockout’) can be used to test whether the corresponding component is required for a certain outcome. Keep in mind however, that a component not required under one set of conditions may still play a role and it may be required under other circumstances. Note also that a knockout animal lacks the component from birth and may compensate for its absence by other biological changes.
Another approach to analysis is depletion of certain immune cells in vivo (often CD4+ or CD8+ T-cells). If animals were intact when primed and are depleted only during the period of viral challenge infection, then depletion is informative about effector functions. It is imperfect in that residual cells could lurk at tissue sites not tested or at levels not detectable. However, depletion has the advantage that it can be performed acutely, not leaving much time for compensation by other changes in the animal.
Multiple approaches are necessary if we are to accumulate a realistic view of immune responses and their potential under various circumstances. No one approach can describe the multifaceted immune response and all its shifting balances.
Immunity to Influenza Virus: B- and T-cell Responses
The immune system clears infection the first time a virus is encountered. It also preserves specific memory of viral antigens, so that it can prevent or at least limit reinfection if the same virus is encountered again. Fundamentals of immune responses (B- and T-cell responses, antigen processing, presentation by major histocompatability complex [MHC] class I and II and epitope dominance) are reviewed elsewhere and will not be covered here.
Antibodies to influenza virus can protect against reinfection and passively transferred antibody can protect naive animals. However, this form of protection is often subtype-specific or even narrowly specific to certain viral strains (Ada and Jones, 1986), failing to protect against mismatched strains (De Jong et al., 2000). Additional immunity is provided by effector T-cells. They play important roles in clearing influenza virus and protecting against challenge, although they can also cause immunopathology (Wells et al., 1981). Doherty’s and other groups have provided much evidence for a beneficial role of class I MHC-restricted CD8+ cytotoxic T-lymphocytes (CTLs) in clearing primary influenza virus infection (Doherty et al., 1997) and also in protection against challenge with homologous virus (Lu and Askonas, 1980). The conserved NP viral protein is a major target antigen for CTLs in mice (Yewdell et al., 1985). In some studies of immunizations with NP, immune responses were observed but little or no protection
(Webster et al., 1991; Lawson et al., 1994), while in other cases NP regimens were protective against challenge (Ulmer et al., 1993; Fu et al., 1999). MHC class II-restricted CTL activity specific for influenza virus antigens has also been reported (Taylor and Bender, 1995).
The role of CD8+ CTLs in protective immunity is virus-specific: CTLs only control the virus they recognize. Bystander viruses coinfecting the same lungs are not controlled (Lukacher et al., 1984), ruling out nonantigen-specific mechanisms based on natural killer (NK) cells or soluble mediators such as interferon (IFN), cytokines, or chemokines released when T-cells recognize virus. Topham and colleagues demonstrated that in vivo protection by CD8+ cells requires lysis mediated by either perforin or Fas (Topham et al., 1997). Tumor necrosis factor (TNF)-α-mediated killing has also been reported in vitro (Liu et al., 1999; Zhao et al., 2001).
Mechanisms of Heterosubtypic Immunity Induced by Infection with Live Virus
Focus will now be placed on Het-I, that is, cross-protection by prior exposure to one influenza A subtype against challenge with a divergent subtype. Respiratory infection with live wild type virus efficiently induces Het-I and will be discussed initially.
Roles of T-cells
T-cells are candidates for contributors to Het-I because they participate in clearing virus from infected tissues and many of them cross-react with all influenza A subtypes. In one study of Het-I, in vivo depletion showed that CD4+ and CD8+ T-cells both contributed to control of challenge virus in the nose (Liang et al., 1994). In the lungs, CD4+ cells did not appear to contribute but CD8+ cells did, plus some other mechanism that remained after depletion of both. This study also showed that Het-I against influenza A was immunologically specific in its effector phase; coinfecting influenza B virus replicated unchecked in the same lung tissue (Liang et al., 1994). Thus, like homologous protection by CTLs discussed earlier, Het-I induced by live virus requires specific effector functions of antibodies or T-cells that recognize the virus.
Mice with a targeted disruption of the β2-microglobulin (β2m) gene have been studied as a model lacking class I MHC restricted CD8+ T-cells. They can survive primary influenza virus infection and can mount protective immune responses to homologous and heterosubtypic challenge (Bender et al., 1994; Epstein et al., 1997). β2m-/- mice have multiple immune deficiencies besides a lack of CTLs, but one can at least say from these results that CD8+ CTLs are not required for Het-I (Raulet, 1994; Epstein et al.,
2000). Confirmatory evidence comes from CD8 knockout mice, which also have deficient class I-restricted CTL and retain Het-I (Nguyen et al., 2001).
Can T-cell responses alone protect against influenza? Immunoglobulin (Ig)-/- knockout mice lacking antibodies and mature B-cells have been used to study this question, including µMT mice (targeted disruption of the membrane exon of µ heavy chain), JHD mice (disruption of the heavy chain joining segments, thus no Ig gene rearrangement) and DI mice (disruption of JHD segments and also κ light chain constant regions). Several studies have shown that such mice could clear primary influenza virus infection but less effectively than normal mice and immunization protected them at least to some extent against homologous challenge (Bot et al., 1996; Topham and Doherty, 1998; Epstein et al., 1998; Graham and Braciale, 1997). What about Het-I? In one study, no Het-I could be demonstrated in µMT mice but under conditions that showed no protection against homologous challenge, either (Nguyen et al., 2001). Indeed, protective immunity is weaker than normal in these mice. Our group has identified conditions under which Het-I could be demonstrated in mice without antibodies. Immunization with H2N2 or H3N2 viruses partially controlled replication of H1N1 challenge virus. This immunity was dependent upon both CD4+ and CD8+ T-cells (Benton et al., 2001).
There is a caveat to interpretation of these results: Ig-/- mice have an immune defect besides absence of antibodies. In a variety of pathogen systems, naive B-cells can restore their ability to clear an infection but not via antibody production (Elkins et al., 1999; Mozdzanowska et al., 2000). These findings suggest a role for B-cells as antigen-presenting cells (APCs).
Role of Antibodies
Antibodies had been suggested as a mediator of Het-I because foster nursing on immune mothers transferred protection to the pups (Mbawuike et al., 1990) and absence of CD8+ CTLs did not abrogate Het-I. What type of antibodies could be involved? IgG dominates in immune serum and reaches mucosal sites, including the lungs by transudation. IgA is found in the lungs along with IgG, and IgA dominates in the nose where it is thought to be especially important. Since mucosal immunization is highly effective, secretory antibodies have been a focus of study and polymeric IgA has been shown to mediate protection against influenza virus (Renegar and Small, 1991). Polymeric IgA can cross the epithelium of the lung and other organs by transcyrosis dependent upon the poly-Ig receptor and can interfere with viral infection as it crosses the infected cells (Mazanec et al., 1992; Mazanec et al., 1995). Certain IgA monoclonal antibodies (mAbs) to core proteins protect against rotavirus infection, although they do not neutralize virus (Burns et al., 1996; Schwartz-Cornil et al., 2002); these results suggested
that during transcytosis antibody interferes with intracellular virus assembly. Antibody to conserved antigens, as required for Het-I, could mediate such a mechanism.
To resolve questions about IgA’s role in control of infections, Harriman and colleagues derived an IgA knockout mouse (Harriman et al., 1999). These mice could clear a primary influenza virus infection and a subunit vaccine protected them against lethal challenge (Mbawuike et al., 1999). We later showed that IgA-/- mice were capable of Het-I under two sets of conditions, lethal challenge infection of the total respiratory tract (TRT) and nonlethal challenge restricted to the upper respiratory tract (URT) (Benton et al., 2001). Thus, Het-I can be effective in the absence of IgA. There may, however, be quantitative defects in control of virus by IgA-/-mice.
Anatomic Compartments and Heterosubtypic Immunity
The influence of routes of immunization on the resulting antiviral responses was demonstrated in 1950 by de St. Groth (de St. Groth and Donnelley, 1950). Immunization of mice with live virus via respiratory tract infection was far more effective than intraperitoneal or subcutaneous immunization in protecting against challenge with homologous virus. In one case, the difference in effectiveness was on the order of a hundred million-fold! The superiority of immunization via mucosal sites presumably reflects viral replication in the respiratory tract and induction of local as well as systemic immunity.
The importance of the anatomic sites in which priming takes place was also highlighted in a study by Nguyen and colleagues. Live virus was given by intraperitoneal or intravenous routes, or by TRT or URT infection (Nguyen et al., 1999). TRT immunization generated better CTL responses than the other routes. It also generated heterosubtypic protection, while the other routes did not. Any of the routes generated cross-reactive CTLs in the spleen and some in the cervical lymph nodes. However, only the TRT route induced CTLs in the mediastinal lymph nodes which drain the lungs and from which CTLs are recruited to the lungs during infection. Depletion of CTLs was not performed to prove they mediated the observed protection, so roles of other local immune responses in Het-I were not ruled out.
Heterosubtypic Immunity Induced by Vaccines Against Influenza A Subtypes Common or Novel in Humans
The previous section explored Het-I induced by wild type virus infection. How can we induce it more safely? This section will explore the ability of various vaccines to induce Het-I. Challenge viruses will include subtypes
in human circulation currently, as well as potential future pandemic subtypes. The studies use different vaccine formulations, virus strains, doses, routes of administration and measures of protection. Since the vaccine candidates have not been compared side by side, general conclusions and comparisons are not yet possible. The details are crucial. This or that preparation may induce cross-protection but how efficiently, at what dose, by what route of administration and against what challenge?
The 1997 outbreak of H5Nl in humans in Hong Kong raised fears that this subtype could spread and perhaps cause a pandemic. Preparation of reassortant vaccine strains was difficult and took over a year (WHO, 2003), while recombinant H5 protein proved not very immunogenic (Nicholson et al., 2001; Treanor et al., 2001). Fortunately, there was no human-to-human transmission and the outbreak subsided. Vaccine candidates face new problems in that pathogenesis of H5N1 infection is different from that of H1N1 and H3N2. H5N1 viruses are extremely virulent in chickens, causing systemic spread, replication in various organs and rapid death (Suarez et al., 1998; Subbarao et al., 1998). Some H5N1 strains spread to nonrespiratory organs in mice and cause symptoms within 24 h and deaths earlier than H1N1 and H3N2 viruses (Gao et al., 1999; Lu et al., 1999). Thus, it was not clear whether immunizations inducing heterosubtypic immunity effective against H1N1 and H3N2 viruses would work against H5N1 infection. Several types of heterosubtypic immunizations have given encouraging results with H5N1 challenge and a few with H9N2 challenge, as will be described in the following sections. These approaches could provide a first-line of defense against a pandemic virus, until antigenically-matched HA-based vaccines could be prepared.
Protein and Peptide Vaccines
A variety of peptide and protein vaccines based on conserved sites confer heterosubtypic protection in animal models, mediated by antibody and/or T-cells. Only a few examples can be mentioned. Despite the generally inefficient entry of exogenous proteins into the class I MHC antigen-presentation pathway, peptides and proteins are able to induce CTL responses to some extent. For example, a fusion protein consisting of part of NS1 fused to the C-terminal part of the HA2 domain of subtype H1 was shown to induce antibody and CTL to influenza virus. It protected mice against challenge with either H1N1 or H2N2 but not H3N2. Depletion of either CD4+ or CD8+ cells partially abrogated this protection (Mbawuike et al., 1994).
Another protein vaccine is based on matrix (M)2. M2 is a conserved protein that spans the virion membrane with a portion exposed on the outside and is a target of protective antibodies. Recombinant M2 protein
has been shown to induce protective immunity against lethal challenge (Slepushkin et al., 1995). Recently Neirynck and colleagues showed that a highly conserved N-terminal peptide of M2 fused to a carrier protein protected mice against homologous and heterosubtypic challenge. This protection was passively transferred by serum antibodies (Neirynck et al., 1999).
Note that vaccines based on peptides or minigenes chosen as dominant T-cell epitopes are unlikely to be effective in responders of all MHC types. Whole genes or proteins providing multiple potential epitopes seem more promising than individual peptides for inducing T-cell immunity in the outbred human population.
DNA Vaccines
Like viral infection, DNA immunization results in endogenous expression of antigens. Thus, in addition to generating antibody responses, it efficiently delivers viral proteins to the antigen-presentation pathways favorable for inducing CTL responses. Extensive studies of DNA vaccination against influenza have used various antigens and routes of administration. Most relevant to Het-I is DNA vaccination with conserved components, such as NP and M. NP DNA induces antibody and T-cell responses and protects against heterosubtypic challenge (Ulmer et al., 1993; Rhodes et al., 1993). Using NP or both NP and M DNA, protection against heterosubtypic challenge was shown to be mediated by both CD4+ and CD8+ cells (Epstein et al., 2000; Ulmer et al., 1998).
In addressing control of infection with H5 viruses, NP DNA gave only modest protection in chickens that was viewed as inadequate (Kodihalli et al., 2000). Nonetheless, we tested the potential of DNA vaccination in mice, using NP and M genes from an H1N1 virus (Epstein et al., 2002). NP + M DNA vaccination protected against lethal challenge with the H5N1 strain HK/156 and reduced lung virus titers approximately 500-fold. NP + M DNA vaccination protected partially against a modest dose of the virulent strain A/HK/483, while DNA vaccination plus viral boosting protected effectively against a higher dose. These results encourage further exploration of DNA vaccination to induce broad cross-protection, with or without viral boosting.
Efforts are being made to improve the potency of DNA vaccination by use of re-engineered plasmids (codon modification to increase expression, optimization of immunostimulatory signals), additional viral antigens, DNA prime-recombinant viral boost strategies, adjuvants and targeting of antigens to favorable antigen presentation pathways. Similar to some other vaccines, DNA vaccines could be prepared in advance and used off the shelf. DNA vaccines have the additional advantage over viruses that they do not require a cold chain. They are currently expensive but if mass
produced, would come down in cost and might become practical for use in parts of the world where the need for refrigeration limits use. Even if DNA vaccination does not turn out to be a method of choice, it provides an analytical technique to test antigenic components separately and optimize the contribution of each to the response.
Inactivated Virus and Subunit Vaccines
Inactivated virus had long been thought relatively ineffective at inducing Het-I, based on studies giving it by systemic routes, such as intramuscularly (Webster and Askonas, 1980). Several recent reports, however, challenge this view and suggest new uses of inactivated vaccines.
In one study (Takada et al., 1999), Takada and colleagues tested as vaccines several inactivated H5 viruses given intranasally. Unexpectedly, a mismatched control virus (inactivated H3N1) given intranasally also protected against lethal H5N1 challenge, suggesting that the new route of administration allowed inactivated vaccine to induce broad protection across a subtype difference.
Tumpey and coworkers studied protection against H5N1 by formalin-inactivated H3N2 virus and analyzed its immune mechanisms (Tumpey et al., 2001). Inactivated virus plus adjuvant given intranasally but not subcutaneously protected mice against lethal H5N1 challenge. The protection was not abrogated by depletion of both CD4+ and CD8+ cells. In addition, the protection was seen in β2µ-/- but not Ig-/- mice. The caveat discussed above, that they have an APC defect, limits the interpretation of the results in the Ig-/- mice. However, in addition, IgG and IgA antibodies reactive with both H3 and H5 HA were detected in the serum and lungs of normal mice immunized intranasally but not subcutaneously. The antibodies neutralized H3N2 but not H5N1 virus. All together, the results suggested a non-neutralizing antibody mechanism of Het-I that is inducible by mucosal but not systemic vaccination with inactivated virus.
These findings were recently extended by Takada and colleagues using inactivated vaccines of HA subtypes H1, H2, H3, H5, and H9 (Takada et al., 2003). When given intranasally, all of these protected mice against challenge with the virulent H5N1 virus, HK/483. IgG antibodies crossreactive with HK/483 viral antigens were detected in the serum and lung wash samples, while IgA was preferentially detected in nasal washes. As in the Tumpey study, antibodies neutralized homologous virus but not H5N1 virus.
Inactivated virus incorporated into ISCOMS (immunostimulating complex [adjuvanted particles]) (Morein et al., 1984) has also been shown to induce Het-I in mice. H1N1 influenza ISCOMS protected mice against lethal challenge with H1N1, H2N2 and H3N2 and reduced lung titers of
H5N1 and H9N2 (Sambhara et al., 2001). In monkeys, flu-ISCOMS failed to protect across the smaller divergence of a drift variant (Rimmelzwaan et al., 2001).
The Takada and Tumpey studies delivered the vaccines under general anesthesia, which would not be a practical method of human vaccination. However, getting antigen to the lungs might be achieved by other means, such as a small particle aerosol. These findings give hope for induction of Het-I with existing vaccines.
Live Attenuated Virus
Use of live attenuated viruses (H5 and H9, for example) as vaccines of potential pandemic subtypes has been proposed by some investigators (Chen et al., 2003; Subbarao et al., 2003; WHO, 2003) but entails some risk. The HA gene of the new subtype could reassort with a nonattenuated virus in the community, leading to release of an infectious, nonattenuated virus of the new subtype. Such an approach would be too risky unless a pandemic were already spreading rapidly in the area and had a high death rate. Since cleavability of HA is related to virulence (Hatta et al., 2001), removing this site by genetic engineering would help by reducing potential pathogenicity.
Live attenuated influenza vaccines of circulating strains (A/H1N1, A/ H3N2 and B) have long been used in Russia, were recently approved for marketing in the USA and have a good safety record. Can live attenuated, cold-adapted vaccines given mucosally induce Het-I and thus protect against potential pandemic subtypes? Recall that wild type virus infection given by TRT exposure in mice induces Het-I but virus infection confined to the URT does not (Nguyen et al., 1999). Cold-adapted vaccine strains infect the URT but replicate poorly in the warmer environment of the lungs of humans or mice. Whether the lower respiratory tract infection they produce is adequate to induce Het-I efficiently is unknown. If they induce Het-I well, these vaccines will not only have a role in controlling annual influenza epidemics but may also have a role in pandemic prevention.
The Possibility of Het-I in Humans
Human immunity to influenza has often been said to be subtype-specific and any heterosubtypic immunity dismissed as ineffective (Murphy and Coelingh, 2002). However, some cross-protection in humans has been reported during consecutive or overlapping epidemics, with reduction both in susceptibility to a second virus and intensity of symptoms (Sonoguchi et al., 1985). During a pandemic, human populations are exposed to an influenza subtype to which they have no prior exposure. If humans have Het-I of
even partial effectiveness and short duration, it could make a major public health contribution.
As discussed above, CTLs are one important mechanism of defense against influenza revealed in the animal studies. Humans have cross-reactive CTLs, too. In human volunteers challenged with H1N1, a cross-reactive memory CTL response correlated with control of infection, even in those individuals with no anti-H1N1 antibody (McMichael et al., 1983a). CTL immunity to influenza in humans has been reported to wane over a period of about 3 to 5 years (McMichael et al., 1983b) but that does not mean we should dismiss its significance.
At the time of the pandemics of 1957 and 1968, enough was known about influenza virus for recorded observations to include virological testing and analysis. The pandemic of 1968 involved a switch only in the HA, so antibodies to NA could have helped control infection. In 1957, however, both the HA and the NA were changed with the switch from H1N1 to H2N2, so this pandemic provides a situation with special potential for analyzing Het-I. In one study of the 1957 pandemic (Slepushkin, 1959), approximately 15,000 workers in a Russian factory were monitored by self-reporting. Influenza-like illness in the spring before emergence of H2N2 gave a 2.2-fold reduction in attacks during the summer when H2N2 began circulating and a 1.6-fold reduction in attacks during the fall. This suggests an impact of prior immunity on susceptibility to the new virus subtype. However, influenza infections were not confirmed by laboratory tests in this study.
Many studies of the 1957 pandemic noted that the incidence of influenza was much higher in children than in adults and declined progressively with age among adults. One active surveillance study, the Cleveland family study, was carried out from 1947 to 1957 and was especially informative for several reasons. The population consisted only of families with young children and thus all the adults were exposed to children as vectors. Furthermore, influenza virus infections were confirmed by culturing of swabs, not just based on symptoms. In this population, the incidence of culture-confirmed influenza was much higher in children than in adults (Jordan et al., 1958). The difference between adults and children was much less pronounced in 1950, 1951 and 1953, during the H1N1 era. This argues against the idea that children are inherently different in some other way, such as behavior or physiology. Thus, the data suggest an effect of prior immunity accumulated over time and exposures. At the time Jordan and colleagues pointed out that serum antibody was not the explanation because sera of their study participants did not inhibit the 1957 virus in HA1 tests (Jordan et al., 1958). They suggested ‘unknown factors’. What has been learned about immunity since then from studies of animal models suggests that
cross-reactive mucosal immunity to viral epitopes conserved between subtypes could be the unknown factor.
If humans have Het-I, we need ways to induce it with safe vaccines. CTL and secretory antibodies are candidates for mediating cross-protection, as in animals. Human CTL recognize a wide variety of influenza antigens, with some epitopes subtype-specific and some cross-reactive (Jameson et al., 1998). Which vaccines induce CTL? Clinical trials of inactivated and live attenuated vaccines, plus the combination of both, are reviewed extensively elsewhere (Couch et al., 1997; Murphy and Coelingh, 2002). Trials are often monitored serologically but the vaccinations that best induce serum antibodies are not always the ones best at inducing CTL or mucosal IgA. Some trials have measured T-cell memory, for example, a trial of inactivated vaccine with and without ISCOMS (Ennis et al., 1999) but clearly some key questions remain to be asked.
Efficacy trials in humans have not been informative about Het-I; they are monitored for prevention of infection with the same subtype(s) as the vaccine, either strain-matched or in some years drift variants. One clinical study raised the question of Het-I in order to see whether it would interfere with subsequent vaccination with a live attenuated virus of a different subtype (Steinhoff et al., 1993). Results showed that prior exposure to wild type virus or cold-adapted vaccine of one influenza A subtype did not interfere with subsequent vaccination with a different cold-adapted vaccine subtype. However, that study was done in young children (6–36 months old). The hints in the historical evidence leave room for the possibility that adults have accumulated a type of immunity that is weaker or more transient in children and thus that Het-I might have potential for public health impact.
The Future: Vaccines and Pandemic Planning
Given the extensive evidence in animals and the hints in humans, the potential of Het-I to reduce morbidity and mortality from a new pandemic strain should be explored. Strainmatched vaccines would probably not be available in time in the case of a rapidly spreading pandemic, even with new technologies for vaccine production. Thus, other anti-infective measures for pandemic intervention are needed, both antiviral drugs and, if continued investigation justifies it, vaccines inducing Het-I. Vaccination strategies using conserved components and experimentally determined to be efficient at inducing Het-I could be used routinely and vaccine could also be offered early in a pandemic to those who had not received it before. It would be intended to offer partial protection as a first line of defense, to be followed by strain-matched vaccines when available. This approach might also be useful to address concern about use of influenza virus as a bioterrorism
threat, because this type of protection does not require prediction of what virus is coming.
An active clinical surveillance study could provide evidence for or against the idea that human Het-I is of a useful magnitude. The study population would be monitored for respiratory illnesses and suspected influenza would be confirmed by viral culture. The vaccination history of participants would also be recorded. Over a period of years, as new viral strains would enter the community, susceptibility could be examined in relation to prior history of influenza virus infections and vaccinations. If a pandemic should occur, the records would be available and the machinery in place to assess the impact of prior infections and vaccinations of different types on susceptibility to the new virus. More likely than a pandemic would be localized outbreaks of novel subtypes (for example, avian viruses transmitted to humans). If such an outbreak occurred in the study population, it might provide an opportunity to evaluate the effects of cross-protection.
Pandemic planning to date has relied mainly on proposals for emergency strain identification, preparation of new vaccines from the pandemic virus and use of antiviral drugs. Additional strategies are required. If heterosubtypic immunity could even partially control infection and thus reduce the morbidity and mortality due to a spreading pandemic, we need to study how best to induce and make use of this type of immunity.
Five-Year View
Many vaccine candidates will be compared in animal models for their ability to protect against potential pandemic subtypes of influenza. Meanwhile, the use of live attenuated vaccines will increase and surveillance of its effectiveness will continue. Besides protecting individuals, this will also increase herd immunity and thus contribute to protection of those who are not vaccinated or who do not respond optimally, such as the elderly. Clinical trials of additional vaccine candidates will assess their potential and, in some cases, the role of immunity to conserved components in protection.
Expert Opinion
Current influenza vaccination practices need improvement, given the high toll of disease. Besides studies of vaccines, clinical surveillance studies are needed to determine the consequences of prior infections and vaccinations in humans as new strains emerge. The potential of heterosubtypic immunity (broad cross-protection) to help control a pandemic has been largely ignored until recently but should be investigated systematically. A large number of vaccine candidates with different advantages and disadvantages are under study. Their ability to induce broad cross-protection
should be one of the elements assessed as they move through clinical trials and into use.
Key Issues
-
Cross-protection against multiple influenza A subtypes can be induced in animals by prior infection or vaccination. Multiple viral antigens and multiple immune effector mechanisms can participate.
-
Mucosal vaccination induces different immune responses than systemic vaccination and is more effective at inducing broad cross-protection to multiple influenza A subtypes in animals.
-
Broad cross-protection in humans is of unclear potency and duration, but epidemiological data suggest that it may have an impact.
-
A variety of vaccines may induce broad cross-protection if administered appropriately.
-
Imperfect vaccine protection is worth having, especially for a virus causing an acute (not latent) infection. It could provide a first line of pandemic defense, to be augmented by subtype- or strain-specific vaccines when available.
Information Resources
-
www.flu.lanl.gov/review/epitopes.html
“Known Influenza Virus Antigenic Peptides Listed by Restricting Major Histocompatibility Complex Molecules,” Suzanne L Epstein, Jonathan W Yewdell, Jack R Bennink.
-
www.cdc.gov/ncidod/diseaseslflu/fluvirus.htm
Centers for Disease Control and Prevention influenza website
-
www.who.int/health-topics/influenza.htm
World Health Organization fact sheet on influenza
GENERATION OF TRANSGENIC CHICKENS RESISTANT TO AVIAN INFLUENZA VIRUS
Laurence Tiley
Department of Veterinary Medicine, University of Cambridge, United Kingdom
and
Helen Sang
Roslin Institute, Midlothian, United Kingdom
Summary
Recent developments in transgenic technologies and inhibitory strategies offer a real opportunity for generating disease-resistant livestock. The domesticated chicken provides an ideal test-bed for investigating the feasibility of achieving this goal in a relevant species. Avian influenza virus presents an attractive target. This disease poses an extremely serious public health threat and is a major economic burden on the poultry industry. Research is underway to determine the most effective transgenic approaches to suppressing influenza virus replication in chickens and to perhaps one day eradicate the disease in this species.
Introduction
Influenza virus is an accomplished species jumper. The natural reservoirs for the virus are aquatic birds such as ducks, geese, and shore birds (Hinshaw et al., 1981; Kawaoka et al., 1988), which contain all of the known subtypes of influenza A. Within these hosts it is usually a relatively benign and genetically stable agent (Webster et al., 1995). Other species are inevitably exposed to these viruses, but usually this does not result in a successful propagative infection. The virus needs to adapt to succeed in the altered environment of a new host. Influenza virus replication readily generates the genetic diversity required for rapid evolution in response to such diversifying selective pressure. Periodically (as in the case of H5N1 in Hong Kong 1997 and subsequent events [Sims et al., 2003]), the virus manages to successfully infect a new host, in this case humans. In common with many emerging viruses, this can result in high case fatality rates among those infected individuals. The crucial adaptation that is needed for full emergence as a new pandemic virus is the ability to transmit efficiently from one individual to another. Fortunately this happened to only a very limited extent with H5N1 in 1997 (Buxton Bridges et al., 2000). However, the three pandemics of human influenza in the 20th century are clear evidence that the virus has succeeded in doing so in the past.
Although in principle it is possible that a new strain of influenza could emerge as a consequence of direct transmission from a wild bird to a human, the evidence suggests that this is extremely unlikely. It is much more likely that an intermediate or bridging host will be involved. Intensively reared domestic livestock such as pigs and poultry are the prime candidates as they are permissive for avian influenza viruses, and can act as amplifier hosts. This results in the massive and prolonged exposure of humans to novel and evolving strains of virus, thus increasing the probability of successful transmission to humans. Reassortment between human and avian strains of virus, either in pigs (which are susceptible to both
[Scholtissek et al., 1985]) or in humans (Claas and Osterhaus, 1998), provides a further means for rapid adaptation to the human host. The level and extent of exposure from infected chickens should not be underestimated. The 2004 epidemic of H5N1 was the largest outbreak of highly pathogenic avian influenza (HPAI) on record and this resulted in at least 34 human cases, 23 of which were fatal. However, there is no reason to expect that the next pandemic strain must be derived from an HPAI. The human pandemic strains H1N1, H2N2, and H3N2 all resemble low-pathogenicity avian influenza (LPAI) viruses. LPAI is prevalent in chickens throughout the world (Alexander, 2003; Senne, 2003).
Nothing can be done to eliminate the small threat of direct transmission from wild birds to humans. However, the major threat posed by the intermediate host species can be tackled in a number of ways, including improved farming/trading practices, vaccination, and biosecurity. A more radical approach, which has now become technically feasible, is to replace these animals with transgenic animals that are resistant to influenza virus. Eliminating avian influenza in chickens internationally would be beneficial on three fronts: It would reduce the risk of cross-species transmission to humans; it would eliminate the economic impact of controlling the disease in poultry; and it would improve the welfare of the animals by reducing the morbidity associated with the disease.
The Time Is Ripe
Basic scientific research into the molecular biology of influenza virus has uncovered several strategies that show great promise for inhibiting influenza replication. Some of these have been known about for many years, but lacked a suitable approach to deliver them.
Mx
The Mx genes were first discovered through their ability to confer a potent antiviral state in mice carrying functional alleles in response to Type I interferons (Haller et al., 1979; Staeheli et al., 1984). In the case of the mouse Mx1 gene, this response is restricted to orthomyxoviruses. Interferon-induced antiviral responses to other viruses (such as Vesicular Stomatitis Virus (VSV) and encephalomyocarditis virus (EMCV)) in mice are independent of Mx1 (Haller et al., 1981). Mx1 alone is both necessary and sufficient to confer this protection, that is, no other IFN-responsive genes are required (Staehli et al., 1986). By contrast, the Mx2 gene of mice is active against VSV and hantaviruses, but inactive against influenza virus (Zurcher et al., 1992). Mx gene homologues have been identified in many other vertebrate species, including humans (Aebi et al., 1989) and avians
(Bernasconi et al., 1995). Human Mx (MxA) has a broad antiviral specificity and is active against influenza virus, measles virus (Schnorr et al., 1993), Semliki Forest virus (Landis et al., 1998), and VSV, among others. The mechanism of action of MxA may be different from that of Mx1, as they have distinct subcellular localizations and antiviral specificities (Dreiding et al., 1985; Pavlovic et al., 1992). For reasons that are not readily apparent, many extensively in-bred lines lack functional Mx genes. This is true for the chicken Mx gene, which has recently been identified (Ko et al., 2002). Mx from most breeds of commercial chickens have a serine amino acid at position 631 and lack antiviral properties whereas functional chicken Mx proteins have asparagine at this position (Ko et al., 2004). The full properties of the chicken Mx gene product have yet to be published, but it appears to follow the MxA pattern of broad efficacy against influenza virus and VSV.
Despite many years of study, the mechanism of action of Mx proteins is still poorly understood. The proteins are all closely related, and all have high levels of homology to dynamin (Obar et al., 1990) and possess consensus GTP-binding domains (Aebi et al., 1989) and a C-terminal leucine zipper (Melen et al., 1992). The broad activity of some Mx proteins suggests an intracellular target intersecting a range of viral pathways. With regard to influenza virus, Mx1 apparently blocks primary transcription of incoming viral genomes (Krug et al., 1985) whereas MxA blocks viral protein synthesis and genome replication, but does not affect primary transcription (Pavlovic et al., 1992). It has been proposed that Mx specifically interacts with the viral polymerase, and evidence exists suggesting this may involve the PB2 (Huang et al., 1992) and/or NP proteins (Turan et al., 2004).
Prospects for Using Mx Genes in Birds
Mouse Mx1 has potent antiviral properties even when transferred into non-murine cell lines lacking endogenous Mx activity (Garber et al., 1991). Chick embryo fibroblast cell lines expressing Mx1 were refractory to influenza virus replication, showing reduced plaquing efficiency, multicycle yield, and viral gene expression. The avian Mx gene inhibits influenza virus and VSV replication when transfected into Mx-negative mouse cell lines (Ko et al., 2004). Either murine or avian Mx genes appear to be promising candidates for introduction into chickens. Restoring Mx function to chickens is likely to be beneficial in controlling influenza, but is unlikely to be sufficient on its own—after all, humans have functional MxA genes, but we still suffer from the flu. This may reflect the fact that Mx is induced by interferon and influenza virus can disrupt interferon responses (one function of the viral NS1 protein [Talon et al., 2000]). Placing the Mx gene under the
control of a constitutive promoter would get around this problem, but may in turn affect the viability of the chicken. Constitutive expression of MxA has been found to promote cell death in response to apoptotic stimuli (Mibayashi et al., 2002). The optimal strategy for expression of Mx needs further investigation.
Decoys (DIs)
RNA decoys are short RNA molecules expressed at high levels that mimic the binding sites for specific RNA-binding proteins. By sequestering these proteins, they act as competitive inhibitors and prevent the normal function of the proteins concerned. Natural decoys exist and are responsible for the formation of defective interfering particles common to certain RNA viruses such as influenza and vesicular stomatitis virus. DI particles are virus particles that contain viral genomes with substantial deletions. By replicating more efficiently than the wild-type virus, they out-compete for host and viral intracellular resources and thus lower the resultant yield of wild-type virus. Natural DIs need to be packaged into virions if they are to persist in cell culture. Therefore, they must contain the signals required for replication and packaging. Synthetic decoys need only to mimic the binding site of the protein. The influenza virus RNA polymerase is a sequence-specific RNA-binding protein complex composed of three viral proteins, PB1, PB2, and PA (Tiley et al., 1994). The complex binds with high affinity to the first 11 bases at the 5′ end of the viral genome (vRNA) (Figure 5-4, boxed sequence). These sequences are absolutely conserved in all segments of all subtypes of influenza A virus (Robertson, 1979) and play several key roles in the control of replication, transcription, and packaging (Li and Palese, 1992; Parvin et al., 1989; Seong and Brownlee, 1992; Hagen et al., 1994; Cianci et al., 1995; Neumann and Hobom, 1995; Lee et al., 2003; Fodor et al., 1994, 1995; Luytjes et al., 1989; Odagiri and Tashiro, 1997). The polymerase also binds to the terminal 10-11 bases at the 5’ end of the complementary replication intermediate (cRNA). These too are extremely well conserved across subtypes.
Experiments in vitro have demonstrated that the interaction of the polymerase with these terminal RNAs is very stable, and decoys based on these sequences have been shown to be potent competitive inhibitors of the polymerase (Luo et al., 1997). Decoy concentration will be a critical factor in the effectiveness of this strategy in vivo. In practice it has been found that the most effective decoys are composed of both the 5′ and 3′ end sequences expressed as a single RNA molecule (so-called panhandle decoys) (Figure 5-4). This may reflect the concentration of decoy achievable in the cell. Panhandle decoys being partially double stranded may be more resistant to intracellular RNAses and thus able to accumulate to higher levels. Alterna-
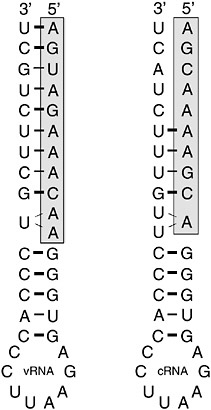
FIGURE 5-4 Typical decoy RNA sequences based upon conserved 5′ and 3′ terminal sequences of influenza virus vRNA and cRNA. Shaded boxes indicate the high affinity polymerase binding site located at the 5′ end of the viral RNAs.
tively, the level of expression provided by the delivery vector alone may be insufficient to achieve significant inhibition. Panhandle decoys contain all the sequences necessary and in the appropriate context for them to be replicated by the influenza virus polymerase. This could result in the decoy levels being amplified by the viral polymerase to levels that impact on virus replication. Clearly this question is crucial to the design of the most efficient decoys. Figure 5-5 shows the effectiveness of a flu-specific RNA decoy in comparison to RNAi-mediated (RNA interference or RNAi) inhibition (see below) using a cell culture-based viral transcription/replication assay. Both approaches can produce substantial levels of inhibition.
Decoy RNAs exploit an interaction between two very highly conserved viral components, the viral polymerase and the terminal sequences of the eight viral genome segments. Thus it is very unlikely that the virus will succeed in circumventing the effect of the decoy by mutation. Should the polymerase mutate such that it has an altered binding specificity, it would
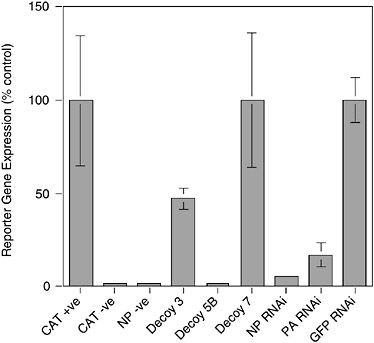
FIGURE 5-5 Inhibition of influenza virus replication by RNA decoys and RNAi. 293T cells were transfected with plasmids expressing influenza virus PB1, PB2, PA, and NP proteins and a plasmid that expresses an RNA corresponding to a negative sense chloramphenicol acetyltransferase (CAT) gene bounded by the 5′ and 3′ termini of vRNA. Transcription of the –ve sense CAT reporter RNA by the influenza virus polymerase leads to the production of CAT protein that can be quantitated by Enzyme Linked Immunosorbent Assay (ELISA). Omission of the CAT reporter (CAT –ve) or polymerase component (NP –ve) ablates CAT gene expression. The effects of various RNA decoy and RNAi expression plasmids are shown. Decoy 3, vRNA sense decoy; Decoy 5B, cRNA sense decoy; Decoy 7, control decoy lacking polymerase binding site; NP and PA RNAi targeting NP and PA mRNA respectively, as reported in Ge et al. (2003); GFP RNAi, irrelevant RNAi targeting the GFP gene.
likely be necessary that each of the termini undergo a compensatory mutation in order to still be recognized. The likelihood of this occurring for all eight segments is very remote.
RNAi
The discovery of the existence of an RNA-mediated gene silencing pathway (RNA interference or RNAi) in a wide range of species, including vertebrates, has already demonstrated enormous potential (for reviews, see
Caplen, 2004; Joost Haasnoot et al., 2003). Short double-stranded RNA molecules 19–21 nucleotides (nt) long, below the threshold for triggering the interferon response, are able to initiate the selective degradation of messenger RNA (mRNA) molecules to which they have the corresponding sequence. Such short interfering RNAs (siRNAs) are unwound and become incorporated into an RNA induced silencing complex (RISC). This complex uses its integral siRNA to recognize and bind to the complementary target mRNA and cleave it in a catalytic reaction using a RISC-associated RNAse. A number of strategies have been developed that allow this system to be exploited using plasmid or viral vector-based approaches. The most versatile so far uses a single strong promoter to drive the expression of a short hairpin RNA (shRNA) that is processed by the cellular protein Dicer into an siRNA suitable for incorporation into the RISC (Paddison et al., 2002). This system mimics the naturally occurring micro RNA (miRNA) system of gene expression control. More sophisticated designs of shRNA modeled closely on the miRNA system are leading to a better understanding of the requirements for effective siRNA molecules (Krol et al., 2004).
RNAi is a catalytic process, and so is less critically dependent on the level of shRNA expression. Design of flu-specific shRNA has concentrated on the highly conserved regions to ensure the RNAis have the broadest range of effectiveness on different subtypes of influenza A virus (Ge et al., 2003). These invariant regions may simply reflect an absence of immune selection, or may be under some other functional constraint of which we are currently unaware. If the former, it is likely that the virus will be able to escape the selective pressure imposed by the RNAi by simply mutating the target site. This has already been demonstrated for RNAi-mediated inhibition of HIV and poliovirus gene expression (Boden et al., 2003; Gitlin et al., 2002). To overcome this it will undoubtedly be necessary to express multiple RNAis against several different targets (akin to combination therapy using antivirals). However, as the number of different siRNAs increases, the efficiency of inhibition by each individual component may decrease as the RISC becomes saturated. This could also have knock-on effects during development. Certain viruses clearly have mechanisms for avoiding RNAi-mediated suppression (Joost Haasnoot et al., 2003). It has been suggested that the influenza virus NS1 protein (an RNA-binding protein itself) is an inhibitor of RNAi (Bucher et al., 2004; Delgadillo et al., 2004; Li et al., 2004). Experimental support for this comes from work on plants and worms, and as yet no evidence for such a function in natural hosts for flu has been reported. RNAi delivered as double-stranded RNA or as shRNA are effective against flu in both cell culture and mice, suggesting that flu is still vulnerable to this approach (Ge et al., 2003; Tompkins et al., 2004).
The Tools Are Now Available
Until very recently, the chief stumbling block to developing influenza-virus resistant chickens was not a lack of ideas, but the lack of a delivery system suitable for engineering the chicken genome. Limited success had been achieved using avian retroviral vectors (Rapp et al., 2003), but the efficiency of transduction was very low, and the levels of transgene expression were poor and not maintained through subsequent generations as a result of transgene silencing. The key breakthrough was the use of lentiviral vectors enveloped in the VSV glycoprotein (G) protein to deliver the transgene package (McGrew et al., 2004). These vectors can be prepared to very high concentrations (1010 transduction units per milliliter [ml]), enter virtually any cell type, and successfully infect and integrate into the chromosomes of cells irrespective of whether these cells are undergoing cell division at the time. For reasons that are still not fully understood, most transgenes introduced by lentiviral vectors do not appear to be subject to gene silencing (Kafri et al., 1997; Naldini et al., 1996).
The efficiency with which lentiviral vectors can transduce the chicken germline has been investigated (McGrew et al., 2004). High-titer preparations of vectors derived from equine infectious anemia virus, pseudotyped with VSV-G, were injected into chick embryos in new laid eggs in ovo, then cultured to hatch. The resulting birds were bred to determine the frequency of production of germline transgenic birds: the frequency achieved using high titers of virus vectors approached 100 percent and the transmission rate of the integrated viruses to the next generation was between 4 percent and 45 percent. The integrated viruses were stably transmitted on to the next generation, suggesting that transgenic lines produced using lentiviral vectors will be stable. Analysis of expression of reporter gene constructs carried by the vectors showed a conserved expression profile between individuals that was maintained after transmission through the germline for at least two generations (Figure 5-6). These results suggest that lentiviral vectors may be used to generate transgenic birds at very high frequencies and that the transgenes carried by the vectors will be expressed in a reliable manner when the transgenic birds are bred. These vectors may be easily engineered to carry siRNA, decoy, and Mx expression constructs. Transgenic mice, produced using lentiviral vectors that carry siRNA expression constructs, have been shown to express effective interfering siRNAs (Rubinson et al., 2003).
Future Hurdles
The first attempts to generate influenza-resistant transgenic chickens are currently underway, and more sophisticated vectors carrying all three of
the inhibitory components described above are in production. However, there is still a long way to go before this research reaches its final objective. The technical objectives are achievable in the relatively short term (3 to 5 years). More challenging will be the need to demonstrate the long-term efficacy, lack of detrimental effects on the chicken or the environment, and safety for human consumption to the satisfaction of the regulatory bodies and the public at large.
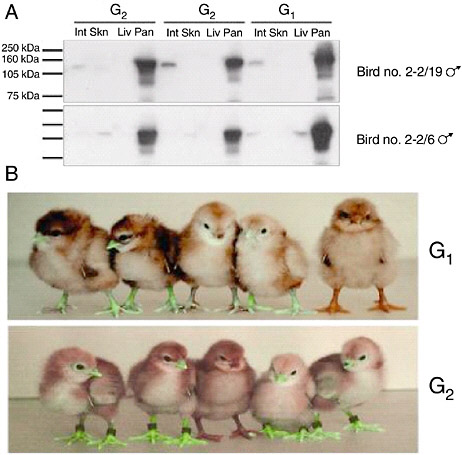
FIGURE 5-6 Reporter gene expression in transgenic birds. A. Western Blot analysis for Lac Z protein extracted from intestine (Int) skin (Skn), liver (Liv), and pancreas (Pan) of G1 cockerels 2-2/19 and 2-2/6 carrying Lac Z expressing lentivector pONY8.0cZ and two G2 offspring from each bird. B. Top panel: five G1 offspring of bird number 4-1. The four birds on the left are transgenic for the green fluorescent protein gene expressed from lentivectors pONY8.0G (all vectors provided by Oxford Biomedica [UK] Ltd). The bird on the right is not transgenic. Lower panel: five G2 offspring from bird number 4-1/66. The bird in the center is nontransgenic.
Is Global Repopulation with Transgenic Chickens Achievable?
Approximately 15 billion chickens are produced each year, so clearly it will take some considerable time, effort, and expense to achieve this goal. For example, approximately sixteen different layer breeds have significant market share worldwide. Each is a three- or four-way cross. Thus, roughly 64 transgenic pure-lines would be required if one wished to reestablish the status quo. Chickens can be bred remarkably quickly. Adult layer stocks produce about 1.8 viable female chicks per week, with an expected viability of more than 90 percent. Replacing the 3 to 4 billion layers would take just 2 to 3 years once the transgenic lines had been made.
Long-Term Efficacy
Transgene expression using lentivectors appears to be very stable, and it is arguable that if transgene expression persists over two generations (as has already been demonstrated in chickens) (McGrew et al., 2004), it is unlikely that it is ever going to suffer from transgene silencing—but time will tell. The selection of resistant mutant viruses is another distinct possibility. By using several independent inhibitory strategies it is hoped that the virus will be unable to overcome the blocks to its replication, thus ensuring the long-term effectiveness of the approach. This is a prerequisite if it is to be worthwhile to move toward the large-scale production of transgenic birds.
Adverse Effects
Because the transgenes are integrated at random in the chicken genome, there is the possibility of deleterious effects in some birds, depending on the location of the integration site. The availability of the chicken genome sequence will make analysis of integration sites quite straightforward, and facilitate the elimination of transgenic birds with the most obviously undesirable gene disruptions. The great advantage of the transgenic approach is that the single desired trait (resistance to influenza virus) can be inserted directly into commercial breeds, obviating the need to introduce the traits by cross-breeding with its associated problems of co-introducing undesirable traits. Once the founder transgenic birds have been fully characterized and shown to be healthy, the birds would be bred normally and subsequent generations would not require repeated genetic modification.
Safety for Humans and the Environment
The strategy used to deliver the transgenes uses a highly efficient and stable delivery vector. The vector has been modified extensively to make it
completely devoid of any viral gene products and it is incapable of replication. The transgenes, decoys, and RNAi are under the control of avian promoter sequences and Mx is a naturally occurring avian gene. ShRNA and decoys are short RNA stemloop sequences that are extremely unlikely to pose any risk to anything other than influenza virus itself. It is difficult to conceive of any realistic risk to human health associated with consumption of such transgenic food. Likewise, chickens carrying such transgenes pose no realistic environmental threat.
Public Attitudes
The prevailing sentiment portrayed by the U.K. media regarding genetically modified products is undeniably negative. However, the majority of the U.K. population are not absolutely against genetically modified organisms (GMO) as food. Most hold the correct view that each GMO must be rigorously assessed on a case-by-case basis. The case for developing influenza-resistant chickens is a strong one on economic, public health, and animal welfare grounds. The risks are extremely small and will be thoroughly assessed. Nevertheless, at least for the foreseeable future in the United Kingdom, there would be significant resistance to the introduction of GMO chickens. Other countries are much more pragmatic about GMO food and are likely to welcome such a development more enthusiastically. Eliminating the chicken from the pandemic influenza equation might delay or prevent the next pandemic disaster. Even the most dire GMO scare-mongering scenario would seem trivial by comparison to a rerun of the Spanish Lady of 1918.
MOLECULAR DIAGNOSTICS IN AN INSECURE WORLD
Michael L. Perdue3
Animal Waste Pathogen Laboratory, Animal and Natural Resources Institute, U.S. Department of Agriculture, Agricultural Research Service
Reprinted with permission, from Perdue (2003) Copyright 2002 by the American Association of Avian Pathologists
Summary
As of October 2001, the potential for use of infectious agents, such as anthrax, as weapons has been firmly established. It has been suggested that attacks on a nation’s agriculture might be a preferred form of terrorism or economic disruption that would not have the attendant stigma of infecting and causing disease in humans. Highly pathogenic avian influenza virus is on every top ten list available for potential agricultural bioweapon agents, generally following foot and mouth disease virus and Newcastle disease virus at or near the top of the list. Rapid detection techniques for bioweapon agents are a critical need for the first-responder community, on a par with vaccine and antiviral development in preventing spread of disease. There are several current approaches for rapid, early responder detection of biological agents including influenza A viruses. There are also several proposed novel approaches in development. The most promising existing approach is real-time fluorescent PCR analysis in a portable format using exquisitely sensitive and specific primers and probes. The potential for reliable and rapid early-responder detection approaches are described, as well as the most promising platforms for using real-time PCR for avian influenza, as well as other potential bioweapon agents.
Current State of Molecular Diagnostics
Homeland defense has become a new item on everyone’s budget request list—including the agricultural world. According to congressional testimonies by D.A. Henderson (2001) and others, rapid detection of introduced biological agents is a critical component in protecting human lives, along with rapid development of vaccines and antimicrobials. While we are all aware that profit margins in poultry production scarcely allow for the kinds of expensive molecular detection equipment that are affordable in the world of human health, the polymerase chain reaction (PCR) as a diagnostic tool has been well established among poultry health professionals for many years (American Association of Avian Pathologists, 1992). Several producers now have their own diagnostic capabilities that include routine PCR analysis for many poultry pathogens. As the market for sophisticated portable detection devices that employ PCR becomes greater, the price of on-site detection for agricultural pathogens will come down. Consequently, it is reasonable to pursue development of detection reagents for high-profile poultry pathogens, particularly the rapidly spreading respiratory pathogens. During an outbreak of a foreign animal disease such as highly pathogenic avian influenza, time is a critical factor in the extent of containment of the disease and the assessment of contamination of surrounding poultry operations and wild bird populations. Fluorescence-based PCR detection can use a single platform for detection of a host of pathogens. Once the
fundamental target genes have been identified and sequenced in the laboratory, the designed primers and probes can be transferred directly to the portable machine format. Many commercially available machines can use the same chemistries, and multiple fluorescent wavelengths should eventually allow multiplex analysis for more than one pathogen per reaction. Also, unlike immunologically based detection methods, fluorescent primers and probes can be altered slightly to accommodate known genetic changes, without having to regenerate and revalidate as with serum-based reagents.
Agricultural Biological Threat Agents
Although agriculture does not immediately come to mind when one considers biowarfare or bioterrorism, every nation that has had a biological warfare program has had an anti-animal and an anticrop component (Rodgers et al., 1999; Wilson et al., 2001). Thus a number of potential threats have been identified for both animals and plants. Expert panels have been convened in recent years to determine attributes most likely to contribute to effectiveness of a bioweapon, and oftentimes the agents have then been ranked. Table 5-2 is a working top ten list of animal pathogens that have been used by the Agricultural Research Service in the last 2 years as a guide for developing detection reagents. At least four of these listed organisms (foot-and-mouth disease [FMD] virus, Newcastle disease virus [NDV], hog cholera virus, and Rinderpest) have been weaponized at one time in the past and evaluated under field conditions (Wilson et al., 2000). Since many of the animal commodity groups, including poultry, are clustered in high concentrations in various regions of the country, the possibility of an event of widespread introduction is high. The reasons for potential purposeful introductions are many and varied, and unfortunately it is not difficult to think of scenarios resulting in purposeful introduction of biological agents into the poultry industry. Thus, we must remain vigilant. The regulatory agencies that would respond to introduction of foreign animal diseases are likely capable of handling a single introduction (Ginsburg, 2000), although the recent FMD outbreak in the United Kingdom clearly illustrates the potential devastating effects of just a single entry point of a highly infectious foreign animal disease. A concerted attack on U.S. poultry with multiple introductions would almost certainly paralyze the industry even with the best efforts of the regulatory agencies.
Avian Influenza A Viruses as Potential Bioweapons
Highly pathogenic avian influenza viruses are generally found on all the lists of potential agricultural bioweapons. Like virulent Newcastle disease viruses that have been weaponized in the past, the AI viruses can be highly
TABLE 5-2 Diseases and Animal Pathogens of Concern to the Agricultural Research Service, U.S. Department of Agriculture
|
Disease |
Agent |
|
Foot and mouth disease |
Apthovirus |
|
Velogenic viscerotropic Newcastle disease |
Paramyxovirus type 1, specific strains |
|
Highly pathogenic avian influenza |
Orthomyxovirus, type A, some subtypes H5 and H7 |
|
Hog cholera |
Pestivirus |
|
Rinderpest |
Morbillivirus |
|
Contagious bovine pleuropneumonia |
Mycoplasma |
|
Lumpy skin disease |
Poxvirus |
|
Blue tongue virus |
Orbivirus |
|
African horse sickness |
Orbivirus |
|
African swine fever |
Asfivirus |
and rapidly infectious via respiratory transmission. Unlike Newcastle disease virus, however, they can further be genetically reassorted in the laboratory to combine genes in a grouping that does not exist in nature, either by mating live viruses or rescuing virus from expression plasmids (Fodor et al., 1999; Hoffmann et al., 2000). The latter plasmid-based techniques are amazing technical developments, but they raise the possibility of major genetic manipulation of viruses and introduction of foreign genes into influenza gene backbones. Consequently, zoonotic influenza viruses in general are getting more attention as potential bioweapons (Peters, 2002). Although it seems highly unlikely that anyone would engineer an avian influenza virus for the purpose of attacking the poultry industry, other potential illegal uses that could spread live or engineered viruses exist. For example, anecdotal evidence exists purporting that poultry farmers have used infectious virus collected from an outbreak to infect their own stock, in attempts to vaccinate or in hopes of indemnification as a result of having infected flocks.
Whatever the case, the need for the capability to rapidly and accurately detect avian influenza viruses, as well as other highly infectious poultry pathogens in the environment, is growing, and new research efforts are needed to evaluate the best approaches to put into the hands of early responders to a purposeful introduction. Availability of validated, rapid, and reliable tests that would supplement the use of slower culture-based detection and immunological subtyping of avian influenza strains would be most useful.
Rapid Detection and Diagnosis
In the face of a scenario where multiple purposeful introductions of avian influenza virus into the poultry industry have occurred, rapid and accurate evaluation of environmental contamination becomes critical, especially if a zoonotic virus is encountered. In such a case the following prioritized attributes of a detection assay would be 1) speed and accuracy, 2) simplicity, 3) common platform for both environmental detection and diagnosis of infected animals, and 4) cost. There are nearly as many approaches to rapidly detecting pathogens and diagnosing disease as there are companies and laboratories developing the technologies. The term diagnostics is widely used to refer to both pinpointing a disease based on the presence of the organism in the host and simply detecting the agent where it should not be. In the veterinary world, generally speaking, the early responders will be state or company veterinarians who will see the disease first, then regulatory agencies that will seek to evaluate the extent of presence or spread. In the case of a zoonotic agent, of course, the public health agencies would become involved quickly. In the case of an agricultural pathogen that does not affect human health, contingency plans are in place to control the spread of disease that depend on the nature of the outbreak. Everyone agrees that the faster the pathogen is detected and the extent of contamination of the environment ascertained, the faster the outbreak can be controlled.
The long-sought, magic, 5-min test for detection is most closely approximated by the antigen capture/enzyme-linked immunosorbent assay (ELISA) based approach, where a sample is loaded onto a filter to which specific antibody is bound and a secondary reporter reagent gives colorimetric verification of presence of an antibody-antigen complex. The Directigen® Flu kit (Becton Dickinson, Franklin Lakes, NJ) is currently being used as a screening tool in disease outbreaks for the detection of any influenza A virus. This technology is rapid and has been used for both humans and poultry for years. It is based on detection of the influenza A nucleoprotein and is not suitable for subtyping strains. The sensitivity of such immunologically based tests is generally lower than nucleic acid based tests, and costs are such that individual bird samples must be pooled before screening.
A number of novel rapid detection approaches employ mass spectrometry (MS) to measure ionization and ion capture profiles following treatment of samples (Donlon and Jackman, 1999). The idea is that environmental samples containing pathogens when treated will yield signature patterns that will instantaneously identify the presence of the pathogen. One of these methods, matrix assisted laser desorption ionization time-of-flight (MALDI-TOF) spectroscopy employs a laser to ionize the sample and
then a measuring chamber to measure the characteristics of the ions as they speed through the chamber. These instruments are continuously being made smaller and smaller, and suitcase size versions are currently being evaluated. Speed of detection is the advantage here if the problems associated with dirty environmental samples can be overcome by preselecting the pathogen out of its native matrix. The procedure will also lend itself to chip-based microarray technology since the laser could move over the array and the time of flight measurements made virtually instantaneously. The future success of such a device in the hands of the early responder will depend on working out the problems associated with preparing the environmental sample for clean measurements by the mass spectrometer. Further, while it is likely that an influenza A virus would provide a characteristic or signature MS profile, the possibilities for obtaining strain-specific characteristics are unknown but would seem very unlikely.
Nucleic-acid based detection and diagnostics will ultimately provide the most information to early responders, scientists, and the regulatory agencies. Just as in DNA forensic analysis in humans, characteristic genetic profiles or gene sequences of microbes can be obtained for each individual species and any strain of that species. Rapid detection of pathogens using PCR amplification of specific genes has been employed for many years, but it requires preparation of an electrophoretic gel run with molecular weight standards to identify and quantify the production of the amplicons. For eukaryotes and DNA containing pathogens, restriction fragment length polymorphisms (RFLPs) and many variations on that idea have been used to unequivocally identify organisms. The same approach can be taken with RNA viruses such as influenza viruses (Offringa et al., 2000), and there are commercially available mobile labs that exploit this approach, such as mobile molecular laboratory (model MML-0150, MJ Research, Waltham MA). Ultimately, small scale, suitcase size nucleic acid sequencers will be available, and this will provide the most information of all to identify environmental pathogens. But this technology is not yet available and is certainly not affordable.
There is one nucleic acid based detection approach for which portable platforms are commercially available and which is becoming accepted by early responders (Fatah, 2001) as a way to identify environmental pathogens, fluorescent real-time/PCR analysis (FRT/PCR). The fundamental chemistry and reaction conditions for this approach have actually been around for years (Heid et al., 1996; Livak et al., 1995) and was originally termed Taqman® PCR (Grove, 1999). Many technological variations of the Taqman chemistry have been developed, including Fret probes and molecular beacon probes, but all use fluorescent probes coupled to PCR for the detection of a wide variety of pathogens. The TAQMAN reaction is extremely specific, more so than other PCR, in that it requires correct
alignment of three separate stretches of sequences in the target pathogen. Like PCR, two primers are designed based on specific target sequences and these amplify a short region of the genome in the regular hot/cool cycle of the PCR. In addition, a probe is designed to bind specifically to the amplicons, and this probe contains a fluorescent molecule on one end and a quencher molecule on the other end. The native exonuclease activity of the polymerase in the PCR cleaves this fluorescent molecule during the reaction releasing its fluorescence from the quenching molecule. So as more amplicon is produced, more probe is bound and more fluorescence is released in real time.
What has happened in the last couple of years is the emergence of commercially available portable machines that can use this technology for a host of pathogens. In addition, the availability of dried or packaged reagents for a variety of pathogens will allow early responders to prepare a sample in the field and run FRT/PCR in place. We have prepared a number of primers and probe reagents that are specific for avian influenza viruses, and these are reported and described by Spackman et al. in this publication.
The advantages of this new technology and reagents are multiple. One is that the reaction is dead end; that is, after the amplification and measurement, the unopened reaction tube is thrown away, reducing the potential for cross contamination. Second, there are now a variety of fluorescent tags of different wavelengths so that the possibility exists for multiplex analysis within the same reaction tube using different wavelength filters to discern positive reactions. Finally, subject matter experts can analyze the reactions in real time in a format that allows for immediate evaluation over the Internet, since web-enabled software is available for some of the portable systems.
Two commercially available portable systems currently exploit these reactions. They are the Idaho technology ruggedized advanced pathogen identification device (RAPID®) system and the Cepheid Corporation SmartCycler®. Benchtop laboratory versions of each are also available from science supply companies. Each portable machine has desirable features that make it different from the other, but rather than go through those here, I refer the reader to the websites for each company: www.idahotech.com and www.cepheid.com. One feature of the RAPID system that makes it particularly attractive is its web-enabled software and potential for wireless transmission that allows transmission of data in real time from the field back to a command center. An epidemiologic tracking system developed in concert with the U.S. Air Force, lightweight epidemiology advanced detection and emergency response (LEADER) uses the RAPID system and allows a central command point to be connected to several machines at once deployed at remote sites at considerable distances apart. So, in terms of potential for early responders, these kinds of systems provide a lot of
promise. With standardized reagents and extraction protocols and communications with a central command post, evaluation of environmental contamination and infection of animals can be quickly evaluated for the regulatory decision makers.
Figure 5-7 illustrates two handheld instruments that use the real-time rapid PCR format and are still in evaluation stages. Efforts continue to miniaturize the detection process such that the technology can be made available to early responders, who would presumably be screening for a single agent to evaluate contamination and spread on the spot. Whether use of these will become a reality in the near future is anyone’s guess. Several hundred of the portable RAPID and SmartCycler machines have been sold to various agencies, such as the National Guard and police departments, but the question is whether dried or prepackaged reagents with long enough shelf lives can be made available. FRT/PCR reagents for other RNA viruses similar to influenza A virus have worked in the prepackaged format, so getting from RNA to real-time analysis is certainly possible. Data presented

FIGURE 5-7 Handheld PCR devices. Two next generation fluorescent real-time PCR devices. On the left is the handheld advanced nucleic acid analyzer (HANAA), a prototype device developed by Lawrence Livermore Laboratories. The device is battery operable and has four cycling chambers to hold PCR reaction tubes. The unit on the right is Idaho Technology’s RAZOR system, which cycles temperatures for the reaction by moving reaction mixtures back and forth in the plastic tubes between fixed temperature chambers and can run 12 reactions simultaneously. In each case, the nucleic acid has to be first extracted from the sample matrix before running the FRT/PCR. With the RAZOR system a set of prepackaged extraction reagents is provided in syringes, and the final sample is injected into the blue plastic receiver tubes. Photos courtesy of Jim Higgins (left) and Idaho Technology (right).
in this symposium have shown that sensitivity of the FRT/PCR approach for avian influenza viruses could be improved, but specificity is very good and costs promise to be cheaper than the immunologically based detection kits. More research is certainly needed to provide prepackaged reagents that could be used by early responders to detect influenza viruses in the environment. However, given the commercial success of this technology, which was used quite extensively during the anthrax attack in the fall of 2001, it is definitely worth pursuing avian influenza specific primer and probe development and validation of assays in real-world settings, such as those described in this symposium.
MODELING PANDEMIC PREPAREDNESS SCENARIOS: HEALTH ECONOMIC IMPLICATIONS OF ENHANCED PANDEMIC VACCINE SUPPLY
Jeroen K. Medema,4,5 York F. Zoellnerb,6 James Ryan,7 Abraham M. Palachea8
Reprinted with permission from Elsevier. Published in Virus Research, Medema et al., 2004. Copyright 2004, Elsevier.
Abstract
Influenza pandemic planning is a complex, multifactorial process, which involves public health authorities, regulatory authorities, academia and industry. It is further complicated by the unpredictability of the time of emergence and severity of the next pandemic and the effectiveness of influenza epidemic interventions. The complexity and uncertainties surrounding pandemic preparedness have so far kept the various stakeholders from joining forces and tackling the problem from its roots. We developed a mathematical model, which shows the tangible consequences of conceptual
|
4 |
Corresponding author: tel: +31-294-477000; fax: +31-294-431164; e-mail: jeroen.medema@solvay.com. |
|
5 |
Business Group Influenza, Solvay Pharmaceuticals BV, P.O. Box 900, Weesp 1380 DA, The Netherlands. |
|
6 |
Department of Health Economics, Solvay Pharmaceuticals GmbH, P.O. Box 220, Hannover 30002, Germany. |
|
7 |
Mapi Values, The Adelphi Mill, Bollington, Macclesfield, Cheshire SK105JB, UK. |
|
8 |
The authors like to thank James Piercy of Mapi Values for the health economic support during development of the model and Professor Kristin L. Nichol of the University of Minnesota Medical School for her valuable comments on the manuscript. |
plans by linking possible pandemic scenarios to health economic outcomes of possible intervention strategies. This model helps to structure the discussion on pandemic preparedness and facilitates the translation of pandemic planning concepts to concrete plans. The case study for which the model has been used shows the current level of global pandemic preparedness in an assumed pandemic scenario, the health economic implications of enhanced pandemic vaccine supply and the importance of cell culture-based influenza vaccine manufacturing technologies as a tool for pandemic control.
Introduction
Influenza is an acute respiratory disease, which can often lead to serious and life-threatening complications in several populations, like the elderly and chronically ill. It occurs in annual epidemics in regions of temperate climates with illness attack rates of 10–20%, leading to an average of 114,000 hospitalizations and 20,000 excess deaths in the United States alone (Strikas et al., 2002). In addition to these annual epidemics, type A influenza viruses have caused pandemics, i.e., sudden global epidemics in all age groups with higher attack and mortality rates. These pandemics are caused by a newly emerging virus subtype in the human population, resulting from reassortment of gene segments between influenza viruses with different host susceptibility (e.g., human, swine and avian strains) or by direct transmission of non-human virus subtypes to humans.
The last century has seen three influenza pandemics, the “Spanish Flu” in 1918–1919, the “Asian Flu” in 1957 and the “Hong Kong Flu” in 1968. The first, one of the most severe events in human history in terms of cases and deaths, was responsible for an estimated 2 billion cases and 20–50 million deaths worldwide (Potter, 1998; Davies, 2000). The latter two were less severe; the 1957 pandemic led to over 1 million deaths worldwide and 70-80,000 in the United States, whereas the “Hong Kong Flu” was reported as relatively mild with peak mortality rates similar to 1957 (Potter, 1998; Strikas et al., 2002). As the influenza virus was only discovered in 1933, influenza diagnosis was poor before that date; hence the corresponding clinical cases have neither been identified nor documented as such before the twentieth century. Analysis of historical documentation, however, clearly indicates that influenza pandemics have occurred at irregular intervals over many centuries and there are no reasons to doubt they will occur again.
The increased awareness of the societal burden of annual influenza epidemics since the mid-1990s led to an increasing awareness of the pandemic threat we are facing. Furthermore, the emergence of two new influenza viruses—subtype H5N1 in 1997 and subtype H9N2 in 1999—in
humans in the Hong Kong area, which seem to be transmitted directly from avian species made this threat real and imminent. The events following 11 September 2001 have further contributed by drawing the general public’s attention to the hazard of biological threats.
The increased awareness on influenza pandemics has led to discussions by public health authorities, regulatory authorities, academia, and industry on what our society can and should do to prepare for the next pandemic. This has resulted amongst others in the founding of the Influenza Vaccine Supply (IVS) Taskforce, a new pandemic discussion and collaboration platform in which all major influenza vaccine manufacturers are represented. Increased awareness has also resulted in governmental pandemic preparedness plans on national and regional levels, such as the “Influenza Pandemic Preparedness Action Plan for the United States” and “A pandemic influenza planning guide for state and local officials” by the Centers of Disease Control and Prevention (Strikas et al., 2002; Patriarca et al., 1999) and the “Canadian contingency plan for pandemic influenza” (Health Canada, 2000). Several attempts are undertaken to bring pandemic planning to multinational or even global level, such as the November 2001 conference on “Preparedness planning in the EU: Influenza and other health threats” and the WHO influenza pandemic preparedness plan (World Health Organization, 1999). Although some regions, e.g. the Canadian province of Ontario, are quite well advanced in pandemic planning and implementation, most of both national and international plans are not completed, let alone realized.
Pandemic planning is a multifactorial process with a consequent high complexity. It is further complicated by the unpredictability of the next pandemic; its time of emergence, spread and severity cannot be foreseen and the efficacy of interventions available for annual influenza epidemics cannot be extrapolated per se to a pandemic situation. Both the complexity and the uncertainties surrounding pandemic preparedness have so far kept the various stakeholders from joining forces and tackling the problem from its roots. The complexity of preparedness planning must be simplified by dividing it into several “sub-projects”, which should be addressed separately to bring pandemic planning from abstract concepts to the desired concreteness.
We developed a mathematical model which shows the tangible consequences of conceptual plans by linking possible pandemic scenarios to health economic outcomes of possible intervention strategies. This model helps to structure the discussion on pandemic preparedness by calculating costs and benefits, gauges the public health benefits of optimized preparedness, and facilitates the translation of pandemic planning concepts to concrete plans.
Materials and Methods
A computer-based simulation model has been developed by MAPI Values (Macclesfield, United Kingdom) and Solvay Pharmaceuticals (Weesp, The Netherlands, and Hannover, Germany), which allows for epidemiological, cost and efficacy inputs as well as manufacturing constraints. The model combines a vaccine production model with an adapted and revised cost-effectiveness model of influenza intervention strategies, based on that developed by Scuffham and West (2002).
Inputs for Pandemic Scenarios
The model inputs for the pandemic scenarios determine the characteristics of the pandemic to which the several intervention strategies can be directed; these input parameters are the attack and mortality rate of that particular pandemic virus, the time of emergence of the pandemic virus and the time for the pandemic to spread and end.
Inputs for Intervention Scenarios
The model inputs for intervention scenarios determine the availability of possible interventions, e.g., pandemic vaccine and antivirals, such as amantadine or the new neuraminidase inhibitors. These input parameters include specific vaccine production inputs, such as time to produce a suitable pandemic virus seed time to produce a batch of vaccine, availability of eggs for egg-based vaccine production and available vaccine manufacturing capacity. They do not include specific antiviral production inputs, as it is assumed that if antivirals are selected as intervention strategy, they will be stockpiled and readily available. Intervention scenario inputs also include parameters, such as pandemic vaccine potency and dosing regime. It is assumed that all manufactured vaccines are used.
Inputs for Intervention Effectiveness
Inputs for intervention effectiveness include parameters such as reduction of influenza-like illness (ILI) cases, reduction in primary care physician (PCP) visits for ILI symptoms, reduction of hospitalizations due to influenza and pneumonia, other respiratory illnesses and congestive heart failure.
Health Economic Inputs
The model allows direct costs, from the perspective of the health care system, to be put in. Parameters for direct costs include all medical costs of
the burden of the pandemic and for the intervention strategy selected. The medical costs for the burden of the pandemic include input parameters like costs of PCP consultations, costs of hospitalization due to influenza and pneumonia, other respiratory illnesses and to congestive heart failure. By combining average costs with resource use (PCP consultations, hospitalizations, etc.), the total medical costs for that particular pandemic can be calculated. The medical costs for the intervention strategy include the vaccine and vaccination cost as well as the costs of antiviral prophylaxis or therapy.
The model allows for a series of secondary input parameters, such as the probability of having an antiviral prescribed during a PCP visit with ILI symptoms, proportion of PCP visits undertaken at home and days off work per case.
Approach
One way of giving structure to conceptual pandemic preparedness plans is to evaluate these plans from a health economic perspective. Therefore the developed model is used to calculate the costs and benefits of certain intervention scenarios for different pandemic scenarios. The strength of the model is its ability to visualize different pandemic scenarios and different preparedness strategies, which is reinforced by assuming the pandemic threat is imminent, rather than taking place at an unknown point in time decades from now. Hence we assumed the next pandemic strain emerges 1 January, 2004, in order to show the current level of pandemic preparedness and how the model can visualize this.
We assumed that the pandemic scenario caused by the “2004 pandemic” virus is based on the 1918–1919 pandemic with influenza attack rates of 20–50% depending on region and a case fatality rate of 1.5% in two pandemic waves (Potter, 1998). For the “2004 pandemic” an average 1918-1919 attack rate of 35% in a single wave with a case fatality rate of 1.87% (Scuffham and West, 2002) are put into the model as pandemic scenario parameters.
Health economic parameters put into the model are based on average data obtained in the elderly in the United Kingdom in recent annual influenza epidemics (Scuffham and West, 2002). For the “2004 pandemic” case study it is assumed that health services utilization during the influenza pandemic are comparable to those during annual epidemics; increased absenteeism of health care workers and societal disruption by the pandemic are not taken into account. It is acknowledged that health care costs and health economical consequences of influenza in the United Kingdom cannot be translated to other countries, especially not to developing countries. Therefore, the latter are left out of this case study and the results that come
from this study only visualize the level of preparedness of the developed countries. The total population of the developed countries is estimated at 1 billion. The key input parameters for the model including vaccination efficacy, are listed in Table 5-3.
Results
The outputs of the model that show the burden of the assumed “2004 pandemic” on developed countries are listed in Table 5-4 under the no intervention scenario. These data show that the no intervention scenario leads to 350 million cases and 22.2, 48.2 and 6.55 million influenza-related PCP consultations, hospitalizations and excess deaths, respectively. Total direct medical care costs add up to € 166.6 billion, with hospitalizations costs of € 165.2 billion as the major contributor.
To visualize the current level of pandemic preparedness, the intervention strategies that can he launched to decrease the burden of the “2004 pandemic” are based on existing egg-based influenza vaccine technologies. Currently, the estimated Northern Hemisphere influenza vaccine usage is 230 million trivalent doses, which translates to a global influenza vaccine manufacturing capacity of 22 million monovalent doses (i.e., 15 µg of pandemic strain antigen) per week. Two main starting materials are needed to manufacture the pandemic vaccine with the current technology: a sufficient amount of embryonated hen’s eggs bred under controlled conditions and a seed of the virus strain to which the vaccine is directed. Preparation of the latter is performed by WHO collaborating centers and takes 1.5–2.5 months for annual epidemic strains. For the assumed “2004 pandemic” seed preparation time is unknown and unpredictable; we assumed a seed preparation time of 3 months, which means that pandemic vaccine manufacture can start 1 April 2004. This is within the regulated annual influenza vaccine manufacturing season, which means that the eggs are readily available for pandemic vaccine manufacture. We assumed that the applied “2004 pandemic” vaccine will be 7.5 µg monovalent given in two doses and should be administered within 6 months after emergence of the “2004 pandemic” virus.
The results of the assumed intervention strategy on the “2004 pandemic” are listed in Table 5-4 as intervention scenario 1. These data show that the current level of pandemic preparedness in the assumed egg-derived scenario leads to vaccination of 17% of the population, which avoids 29.8 million influenza cases, 1.74 million PCP visits, 267 million hospitalizations and 556,000 deaths compared to a scenario of no vaccination. The intervention strategy costs add up to € 2.2 billion, but as this strategy leads to a reduction of € 9.3 billion medical care costs, the saving on direct costs of the “2004 pandemic” is € 7.1 billion.
TABLE 5-3 Key Input Parameters Used Within the Model
|
Vaccine production |
|
|
Month of pandemic strain |
|
|
Month egg seed released by WHO |
April 2004a |
|
Month last vaccine required in market |
|
|
Number of weeks to produce a vaccine |
|
|
Weeks cell seed released before egg seed |
9b |
|
Size of (trivalent) vaccine unit (µg) |
|
|
Number of shots required per person |
|
|
Dose per shot (µg) |
|
|
Annual manufacturing capacity |
|
|
Demographics |
|
|
Overall population |
|
|
Target population |
|
|
Influenza attack rate (%) |
|
|
Years per life lost |
5c |
|
Event probabilities |
|
|
PCP consultations (for influenza) |
6.34d |
|
Hospitalization rates due to: |
|
|
Influenza + pneumonia |
2.52d |
|
Other respiratory disease |
9.34d |
|
Congestive heart failure |
1.9d |
|
Mortality rate |
1.87d |
|
Vaccine effectiveness |
|
|
Vaccine efficacy (%) |
53e |
|
Reduction in GP visits (%) |
46f |
|
Reduction in hospitalizations due to: |
|
|
Influenza + pneumonia (%) |
39f |
|
Other respiratory disease (%) |
32f |
|
Congestive heart failure (%) |
27f |
|
Reduction in mortality (%) |
50f |
|
Unit costs and economic parameters |
|
|
PCP consultation (€) |
28.6c |
|
Influenza vaccine price (€) |
|
|
Influenza vaccine service and administration(€) |
3.1c |
|
Discount rate |
5c |
|
Hospitalization for: |
|
|
Influenza + pneumonia (€) |
3,585c |
|
Other respiratory disease (€) |
3,362c |
|
Congestive heart failure (€) |
3,556c |
|
Average hospital cost per bed per day (€) |
261c |
|
aAuthors’ assumption for egg-based vaccine intervention strategies. bAuthors’ assumption for cell culture-based vaccine intervention strategies. cScuffham and West (2002). dDerived from Scuffham and West (2002). eGovaert et al. (1994). fNichol et al. (1998). |
|
TABLE 5-4 Burden of Different Intervention Scenarios in the Assumed Pandemic
|
|
No Intervention |
Intervention Scenario 1 Egg-Based Vaccine Manufacture |
Intervention Scenario 2 Cell Culture-Based Vaccine Manufacture |
|
Vaccination coverage (%) |
0 |
17 |
37 |
|
Number vaccinated |
0 |
170,000,000 |
370,000,000 |
|
Number of cases (influenza) |
350,000,000 |
320,250,000 |
285,250,000 |
|
PCP visits for treatment (influenza) |
22,190,000 |
20,454,742 |
18,413,262 |
|
Hospitalizations |
48,160,000 |
45,491,663 |
42,352,443 |
|
Excess deaths |
6,545,000 |
5,988,675 |
5,334,175 |
|
Discounted years per life lost |
25,640,894 |
23,461,418 |
20,897,328 |
|
PCP visit costs vaccinations (€) |
0 |
2,227,000,000 |
4,847,000,000 |
|
PCP visit costs treatment (€) |
1,007,181,910 |
928,420,285 |
835,759,549 |
|
Hospitalization costs (€) |
165,170,880,000 |
156,010,312,598 |
145,233,174,478 |
|
Strategy costs (€) |
0 |
2,227,000,000 |
4,847,000,000 |
|
Medical care costs (€) |
166,647,295,200 |
157,371,272,130 |
146,458,303,811 |
|
Total direct costs (€) |
166,647,295,200 |
159,598,272,130 |
151,305,303,811 |
The question for pandemic planners is if the level of pandemic preparedness can be improved and which intervention strategies should be selected to achieve this. As vaccination is the most cost effective intervention, an improved intervention strategy from a health economic perspective would be to produce more pandemic vaccine in the available time frame. This can be achieved by increasing the time period for vaccine manufacture, by increasing the vaccine manufacturing capacity or preferably by a combination of the two.
An earlier availability of a suitable seed virus would increase the time frame for pandemic vaccine manufacture and hence improve the intervention strategy. The new cell culture-based influenza vaccine manufacturing technology likely facilitates seed preparation, as mammalian cell culture may require less adaptation of viruses and shows containment of more pathogenic pandemic viruses without prior decrease of their virulence (see Section 49). To visualize this we calculated the level of pandemic preparedness with an intervention scenario of cell culture-based vaccine manufacturing, assuming that preparation of a cell culture-based seed virus takes 1 month and assuming that the current global weekly manufacturing capacity of 22 million monovalent doses is cell culture-based instead of egg-based. The results of this assumed intervention strategy on the “2004 pandemic” are listed in Table 5-4 as intervention scenario 2.
These results show that, compared to no intervention scenario, the cell culture-based intervention strategy avoids 75 million influenza cases, 3.78 million PCP consultations for influenza treatment and, respectively, 5.81 million and 1.21 million influenza-related hospitalizations and excess deaths. Compared to the assumed egg-based vaccine intervention with 17% vaccine coverage, the cell culture-based intervention strategy leads to vaccination of 37% of the population, avoiding an additional 35 million influenza cases, 2.04 million PCP consultations for influenza treatment, 3.14 million influenza-related hospitalizations and 654,500 excess deaths. The cell culture-based intervention strategy costs add up to € 4.9 billion, but as this strategy leads to a reduction of € 20.2 billion medical care costs, the saving on direct costs of the “2004 pandemic” is € 15.3 billion, an additional € 8.3 billion compared to the egg-based intervention scenario.
The years per life lost gained are € 2.56 million, whereas the cost per life year gained is € 3198. The cost per case, hospitalization and death averted are, respectively, € 234, € 2612 and € 12,530.
Discussion
The “2004 pandemic” case study is based on numerous assumptions. Each can be challenged and, indeed, such uncertain variables are discussed by pandemic planners to challenge preparedness concepts. However, the assumed parameters are only inputs to the presented model and not part of the model itself. Therefore, questioning these assumptions is not questioning the model itself. The model is only a tool to visualize the consequences and effectiveness of a number of planning concepts, to which it lends struc-
ture and logic. Moreover, a sensitivity analysis of the model can be used to set priorities within pandemic planning discussions: the more sensitive the model is to a given assumption, the higher the effect of that particular input on the level of pandemic preparedness.
A sensitivity analysis of the “2004 pandemic” case study shows that the model is most sensitive to the pandemic scenario in terms of attack and mortality rates, as well as to the intervention scenarios in terms of effectiveness and available amounts of pandemic vaccine. It is less sensitive to medical care costs and relatively insensitive to pandemic vaccine price. The total direct medical costs of € 166.6 billion are remarkably close to the € 166.5 billion calculated by the CDC for a pandemic with 35% attack rate (Meltzer et al., 1999). However, these are for the United States alone with 89 million cases, 734,000 hospitalizations and 207,000 excess deaths on a smaller population than in the model presented here. Although the CDC model is similar, the inputs are based on U.S. data, explaining the higher costs.
Pandemic Scenario
The assumptions put into the model to derive the presented “2004 pandemic” scenario have been based on the 1918–1919 pandemic in terms of spread and severity. The first cases were reported in March 1918, leading to a first wave that was not severe. It took about 5–6 months before the pandemic evolved into a second wave with the particular characteristics of high attack and mortality rates, in the age groups at risk for epidemic influenza as well as other age groups, like children and young adults (Potter, 1998). This pandemic shows that everybody can be at risk for pandemic influenza and means that at that time intervention in all age groups at risk should have taken place in these 6 months between emergence and actuality of the pandemic. However, both knowledge and surveillance of circulating influenza viruses have been well established over the last 50 years, enabling an earlier identification of potential pandemic viruses and consequently an earlier onset of intervention programs compared to 1918. Furthermore, improved influenza diagnosis and the availability of intervention enable us to significantly reduce pandemic virus spread. On the other hand, the increased globalization and international travel in current times are likely to speed up spread of pandemic viruses to a large extent. As these phenomena counteract, we assumed for the “2004 pandemic” the same time frame between emergence and spread of the pandemic virus as in 1918.
The severity of a pandemic is reflected in the model by attack rate and event probabilities, such as influenza-related PCP consultations, hospitalizations and case fatality rate. For the “2004 pandemic” we assumed a higher attack rate to reflect a higher severity compared to annual epidemics. All event probabilities for the “2004 pandemic” are based on annual epi-
demic UK event probabilities for the elderly, which are higher than for other age groups. However, the higher attack rate implies that during the “2004 pandemic” other age groups are affected as well and extrapolation of high event probabilities in the elderly to these age groups also reflects a more severe event.
Egg-Based Vaccine Intervention Scenario
Health economic studies on annual influenza epidemics have shown that influenza vaccination is by far more cost-effective than prescription of antivirals for prophylaxis or therapy (Scuffham and West, 2002). Because the health economic perspective has been used in the model to visualize the current level of pandemic preparedness, vaccination has been selected as the pandemic intervention strategy in the current case study as well. The case study is limited to the developed world, because the health economic inputs are based on data gathered in a developed country, but pandemics are a global event and the burden in developing countries might be even greater because of lower public health. However, as long as global pandemic preparedness plans have not been realized, the world will face a limited availability of interventions. These will be made available to developed countries first at the cost of the developing world.
The intervention scenario input parameters are based on current influenza vaccine manufacturing technologies. We assumed that the applied “2004 pandemic” vaccine will be 7.5 µg monovalent given in two doses, as the human population is immunologically naive to the pandemic strain and will need a booster-dose to elicit a protective immune response. Clinical studies in unprimed individuals indeed indicate that a two dose regime is needed (Nicholson et al., 1979), but that this may be achieved with a lower, possibly adjuvanted dose (Nicholson et al., 2001; Hehme et al., 2002).
The next pandemic is likely to be detected by the WHO global influenza surveillance network and, just as for annual epidemic strains, WHO collaboration centers prepare the pandemic virus seed. Seed preparation time for the next pandemic is unpredictable and cannot be based on experience with epidemic strains: although the H5N1-subtype that emerged in Hong Kong in 1997 appeared only a candidate pandemic virus, a suitable seed is still not available 5 years later. In case of a high pandemic threat the efforts that will be put into seed preparation will be accordingly and preparation times for pandemic seeds suitable for egg-based vaccine manufacture in general is expected to be 2–8 months (internal communication J. Woods of WHO Collaboration Center NIBSC, WHO Experts-IVS Taskforce Meeting, September 26, 2002, Geneva). For the case study we assumed 3 months, leaving 3 months for vaccine manufacture, distribution and vaccination before the “2004 pandemic” would evolve and widely spread.
Field strains circulating in the human population usually do not propagate well on eggs and need to be attenuated for egg-based vaccine manufacture. Therefore, both epidemic and pandemic seed preparation for egg-based technology mainly consists of preparation of so-called high growth reassortants by reassortment of field strains with strains that propagate well on eggs. Additionally, pandemic viruses are more virulent than epidemic strains and need to be attenuated to decrease their virulence to safely propagate such viruses in existing egg-based manufacturing facilities, as these are based on open systems.
On the contrary, cell culture-based vaccine manufacture is performed in closed bioreactor systems, enabling the desired containment to process more virulent pandemic viruses without prior attenuation and hence less laborious seed preparation. Moreover, cell culture-based influenza vaccine manufacturing technologies make use of mammalian cell lines, such as MDCK, which are more closely related to the human host than the avian egg-based technology. MDCK-grown human influenza field isolates indeed are generally more antigenically homogenous and more alike the original field strain than egg-grown isolates (Zambon, 1998), which is probably caused by host cell selection of variants. Mammalian epithelioid cells like MDCK appear to be the most sensitive cell culture system for human influenza viruses to date (Zambon, 1998), whereas some human influenza isolates need to be adapted to growth in the allantoic cavity (Murphy and Webster, 1996). Therefore, cell culture is generally used for primary isolation of human influenza viruses. The probability that human pandemic viruses readily grow in mammalian cells therefore seems higher than in the more distant avian host, thereby making the preparation of high growth reassortants obsolete and decreasing the time for pandemic seed preparation. This favors the preparation of pandemic seeds by propagation of the pandemic strain directly on mammalian cell culture instead of via embryonated eggs. However, for annual epidemic vaccine manufacture the egg-passage is a regulatory obligation; omission of this procedure therefore needs to be cleared by authorities.
As the time period for vaccine manufacture is mainly determined by the time a suitable virus seed is available and the time the pandemic spreads in the population, a potential quicker release of pandemic seed suitable for cell culture enables the production of more vaccines. The more vaccines, the greater the vaccination rate and the greater the opportunity to benefit from both reduced mortality and reduced total costs. This underlines the importance of cell culture-based influenza vaccine manufacturing as tool for increased pandemic preparedness.
By assuming the pandemic virus emerges in January, eggs are readily available for pandemic vaccine manufacture once the seed is available. However, a pandemic virus can emerge all year round, also outside the
annual epidemic vaccine manufacturing period. Given that logistics of egg preparation require ordering about 1 year in advance, availability of sufficient eggs outside the planned period certainly can be questioned. Cell culture-based vaccine manufacture however makes long-term advance planning obsolete, as all starting materials are in stock and readily available whenever required. This advantage however is not illustrated in the assumed “2004 pandemic”, thereby underestimating the advantageous health economics of cell culture-based over egg-based intervention and thus the importance of cell culture-based vaccine manufacturing as tool for increased pandemic preparedness.
Although the presented model can lend structure and logic to pandemic preparedness discussions, pandemic planning remains a complex process. The efficacy of interventions, such as vaccines and antivirals, during pandemics needs to be studied and proven beforehand as much as possible in order to make such interventions available in time. Regulatory procedures of such interventions need to be harmonized and adapted to the short-time lines available in pandemic situations and logistical systems need to be set up or streamlined to successfully execute the intervention strategy. Pandemic vaccination might be the most cost-effective approach, but will require the availability of a suitable virus seed and adequate manufacturing facilities to process such a seed. In order to manufacture sufficient amounts for an adequate level of pandemic vaccine, manufacturing capacity needs to be increased. As stated by WHO, policy makers need to keep in mind the several years needed to construct new production facilities and significantly increase production capacity (World Health Organization, 2002). This only has an economical incentive if interpandemic vaccine usage is increased, which—just as pandemic preparedness—is a joint responsibility of the public and private sector.
REFERENCES
Ada GL, Jones PD. 1986. The immune response to influenza infection. Curr Top Microbiol Immunol 128:1–54.
Aebi M, Fah J, Hurt N, Samuel CE, Thomis D, Bazzigher L, Pavlovic J, Haller O, Staeheli P. 1989. cDNA structures and regulation of two interferon-induced human Mx proteins. Mol Cell Biol 9:5062–5072.
Alexander DJ. 2003. Report on avian influenza in the Eastern Hemisphere during 1997–2002. Avian Diseases 47:792–797.
Altmuller A, Fitch WM, Scholtissek C. 1989. Biological and genetic evolution of the nucleoprotein gene of human influenza A viruses. J Gen Virol 70:2111–2119.
American Association of Avian Pathologists. 1992 (August). Proceedings of the Symposium on Biotechnology Applications in Avian Medicine. American Veterinary Medical Association Meeting, Boston, MA.
Bender BS, Bell WE, Taylor S, Small PA Jr. 1994. Class I major histocompatibility complex-restricted cytotoxic T lymphocytes are not necessary for heterotypic immunity to influenza. J Infect Dis 170(5):1195-1200.
Benton KA, Misplon JA, La C-Y, Bruckiewicz RR, Prasad SA, Epstein SL. 2001. Heterosubtypic immunity to influenza A virus in mice lacking either IgA, all Ig, NKT cells, or gd T-cells. J Immunol 166:7437–7445.
Bernasconi D, Schultz U, Staeheli P. 1995. The interferon-induced Mx protein of chickens lacks antiviral activity. J Interferon Cytokine Res 15:47–53.
Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. 2003. Human immunodeficiency virus type 1 escape from RNA interference. J Virol 77:11531–11535.
Bot A, Reichlin A, Isobe H, Bot S, Schulman J, Yokoyama WM, Bona CA. 1996. Cellular mechanisms involved in protection and recovery from influenza virus infection in immunodeficient mice . J Virol 70:5668–5672.
Bucher E, Hemmes H, de Haan P, Goldbach R, Prins M. 2004. The influenza A virus NS1 protein binds small interfering RNAs and suppresses RNA silencing in plants. J Gen Virol 85:983–991.
Burns JW, Siadat-Pajouh M, Krishnaney AA, Greenberg HB. 1996. Protective effect of rotavirus VP6-specific 19A monoclonal antibodies that lack neutralizing activity. Science 272:104–107.
Buxton Bridges C, Katz JM, Seto WH, Chan PK, Tsang D, Ho W, Mak KH, Lim W, Tam JS, Clarke M, Williams SG, Mounts AW, Bresee JS, Conn LA, Rowe T, Hu-Primmer J, Abernathy RA, Lu X, Cox NJ, Fukuda K. 2000. Risk of influenza A (H5N1) infection among health care workers exposed to patients with influenza A (H5N1), Hong Kong. J Infect Dis 181:344–348.
Caplen NJ. 2004. Gene therapy progress and prospects. Downregulating gene expression: The impact of RNA interference. Gene Ther 11:1241–1248.
CDC (Centers for Disease Control and Prevention). 2000. Updated recommendations from the Advisory Committee on Immunization Practices in response to delays in supply of influenza vaccine for the 2000–01 season. MMWR 49:888–892.
Chen HL, Subbarao K, Swayne D, Chen Q, Lu X, Katz J, Cox N, Matsuoka Y. 2003. Generation and evaluation of a high-growth reassortant H9N2 influenza A virus as a pandemic vaccine candidate. Vaccine 21:1974–1979.
Cianci C, Tiley L, Krystal M. 1995. Differential activation of the influenza virus polymerase via template RNA binding. J Virol 69:3995–3999.
Claas EC, Osterhaus AD. 1998. New clues to the emergence of flu pandemics. Nat Med 4:1122–1123.
Claas EC, Osterhaus AD, van Beek R, De Jong JC, Rimmelzwaan GF, Senne DA, Krauss S, Shortridge KF, Webster RG. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472–477.
Couch RB, Keitel WA, Cate TR. 1997. Improvement of inactivated influenza virus vaccines. J Infect Dis 176(Suppl 1):S38–S44.
Davies P. 2000. Catching Cold: 1918s Forgotten Tragedy and the Scientific Hunt for the Virus That Caused It. Harmondsworth, UK: Penguin Books.
de Jong JC, Beyer WE, Palache AM, Rimmelzwaan GF, Osterhaus AD. 2000. Mismatch between the 1997/1998 influenza vaccine and the major epidemic A(H3N2) virus strain as the cause of an inadequate vaccine-induced antibody response to this strain in the elderly. Med Viral 61:94–99.
de St. Groth SF, Donnelley M. 1950. Studies in experimental immunology of influenza. IV. The protective value of active immunization. Aust J Exp Biol Med Sci 28:61–75.
Delgadillo MO, Saenz P, Salvador B, Garcia JA, Simon-Mateo C. 2004. Human influenza virus NS1 protein enhances viral pathogenicity and acts as an RNA silencing suppressor in plants. J Gen Virol 85:993–999.
Doherty PC, Topham DJ, Tripp RA, Cardin RD, Brooks JW, Stevenson PG. 1997. Effector CD4(+) and CD8(+) T-cell mechanisms in the control of respiratory virus infections. Immunol Rev 159:105–117.
Donlon M, Jackman J. 1999. DARPA integrated chemical and biological detection system. Johns Hopkins APL Technical Digest 20:320–325.
Dreiding P, Staeheli P, Haller O. 1985. Interferon-induced protein Mx accumulates in nuclei of mouse cells expressing resistance to influenza viruses. Virology 140:192–196.
Elkins KL, Bosio CM, Rhinehart-Jones TR. 1999. Importance of B-cells but not specific antibodies, in primary and secondary protective immunity to the intracellular bacterium Franciella tularensis live vaccine strain. Infect Immun 67:6002–6007.
Ennis FA, Cruz J, Jameson J, Klein M, Burt D, Thipphawong J. 1999. Augmentation of human influenza A virus-specific cytotoxic T-lymphocyte memory by influenza vaccine and adjuvanted carriers (ISCOMS). Virology 259:256–261.
Epstein SL. 2003. Control of influenza virus infection by immunity to conserved viral features. Expert Rev Anti Infect Ther 1(4):627–638.
Epstein SL, La CY, Misplon JA, Lawson CM, Hendrickson BA, Max EE, Subbarao K. 1997. Mechanisms of heterosubtypic immunity to lethal influenza A virus infection in fully immunocompetent, T cell-depleted, beta2-microglobulin-deficient, and J chain-deficient mice. J Immunol 158:1222–1230.
Epstein SL, La CY, Misplon JA, Bennink JR. 1998. Mechanism of protective immunity against influenza virus infection in mice without antibodies. J Immunol 160:322–327.
Epstein SL, Stack A, Misplon JA, Lo CY, Mostowski H, Bennink J, Subbarao K. 2000. Vaccination with DNA encoding internal proteins of influenza virus does not require CD8(+) cytotoxic T lymphocytes: Either CD4(+) or CD8(+) T cells can promote survival and recovery after challenge. Intl Immunol 12:91–101.
Epstein SL, Tumpey TM, Misplon JA, Lo CY, Cooper LA, Subbarao K, Renshaw M, Sambhara S, Katz JM. 2002. DNA vaccine expressing conserved influenza virus proteins protective against H5N1 challenge infection in mice. Emerg Infect Dis 8:796–801.
Fatah AA. 2001. An introduction to biological agent detection equipment for emergency first responders. U.S. Department of Justice, National Institute of Justice Publication NIJ Guide.
Fodor E, Pritlove DC, Brownlee GG. 1994. The influenza virus panhandle is involved in the initiation of transcription. J Virol 68:4092–4096.
Fodor E, Pritlove DC, Brownlee GG. 1995. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J Virol 69:4012–4019.
Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, Garcia-Sastre A. 1999. Rescue of influenza A virus from recombinant DNA. J Virol 73:9679–9682.
Fu TM, Guan LM, Friedman A, Schofield TL, Ulmer JB, Liu MA, Donnelly JJ. 1999. Dose dependence of CTL precursor frequency induced by a DNA vaccine and correlation with protective immunity against influenza virus challenge . J Immunol 162:4163–4170.
Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley AJ, Kawaoka Y. 1999. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol 73:3184–3189.
Garber EA, Chute HT, Condra JH, Gotlib L, Colonno RJ, Smith RG. 1991. Avian cells expressing the murine Mx1 protein are resistant to influenza virus infection. Virology 180:754–762.
Ge Q, McManus MT, Nguyen T, Shen CH, Sharp PA, Eisen HN, Chen J. 2003. RNA interference of influenza virus production by directly targeting mRNA for degradation and indirectly inhibiting all viral RNA transcription. Proc Natl Acad Sci USA 100:2718–2723.
Ginsburg J. 2000, September 11. Bioinvasion. Business Week. Pp. 70–78.
Gitlin L, Karelsky S, Andino R. 2002. Short interfering RNA confers intracellular antiviral immunity in human cells. Nature 418:430–434.
Glezen WP, Six HR, Perrotta DM, Decker M, Joseph S. 1984. Epidemics and their causative viruses—Community Experience. In: Stuart Harris CH, Potter CW, eds. The Molecular Virology and Epidemiology of Influenza. London, England: Academic Press. Pp. 17–32.
Govaert TM, Sprenger MJ, Dinang GJ, Aretz K, Masurel N, Knotternus JA. 1994. Immune response to influenza vaccination of elderly people. A randomized double-blind placebo-controlled trial. Vaccine 12:1185–1189.
Graham MB, Braciale TJ. 1997. Resistance to and recovery from lethal influenza virus infection in B-lymphocyte-deficient mice. Exp Med 186:2063–2068.
Grove DS. 1999. Quantitative real-time polymerase chain reaction for the core facility using TaqMan and the Perkin-Elmer/Applied Biosystems Division 7700 Sequence detector. J Biomol Tech 10:11–16.
Hagen M, Chung TD, Butcher JA, Krystal M. 1994. Recombinant influenza virus polymerase: Requirement of both 5' and 3' viral ends for endonuclease activity. J Virol 68:1509–1515.
Haller O, Arnheiter H, Gresser I, Lindenmann J. 1979. Genetically determined, interferon-dependent resistance to influenza virus in mice. J Exp Med 149:601–612.
Haller O, Arnheiter H, Gresser I, Lindenmann J. 1981. Virus-specific interferon action. Protection of newborn Mx carriers against lethal infection with influenza virus. J Exp Med 154:199–203.
Harriman GR, Bogue M, Rogers P, Finegold M, Pacheco S, Bradley A, Zhang Y, Mbawuike IN. 1999. Targeted deletion of the IgA constant region in mice leads to IgA deficiency with alterations in expression of other Ig isotypes. J Immunol 162:2521–2529.
Hatta M, Gao P, Halfmann P, Kawaoka Y. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840–1842.
Health Canada. 2000 (January 5). Canadian Contingency Plan for Pandemic Influenza—Draft. The Laboratory Center for Disease Control, Health Canada, Ottowa.
Hehme N, Engelmann H, Kunzel W, Neumeier E, Sanger R. 2002. Pandemic preparedness lessons learnt from H2N2 and H9N2 candidate vaccines. Med Microbio Immunol 191:203–208.
Heid CA, Stevens J, Livak KJ, Williams PM. 1996. Real time quantitative PCR. Genome Res 6:986–994.
Henderson DA. 2001. Hearing on the Threat of Bioterrorism and the Spread of Infectious Diseases. Statement at the September 5, 2001, hearing of the Committee on Foreign Relations, U.S. Senate. [Online]. Available: http://www.hopkins-biodefense.org/pages/library/spread.html.
Hinshaw VS, Webster RG, Rodriguez RJ. 1981. Influenza A viruses: Combinations of hemagglutinin and neuraminidase subtypes isolated from animals and other sources. Arch Virol 67:191–201.
Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA 97:6108–6113.
Huang T, Pavlovic J, Staeheli P, Krystal M. 1992. Overexpression of the influenza virus polymerase can titrate out inhibition by the murine Mx1 protein. J Virol 66:4154–4160.
Jameson J, Cruz J, Ennis FA. 1998. Human cytotoxic T-lymphocyte repertoire to influenza A viruses. Virology 72:8682–8689.
Joost Haasnoot PC, Cupac D, Berkhout B. 2003. Inhibition of virus replication by RNA interference. J Biomed Sci 10:607–616.
Jordan WS Jr, Denny FW Jr, Badger GF, Curtiss C, Dingle JH, Oseasohn R, Stevens DA. 1958. A study of illness in a group of Cleveland families. XVII. The occurrence of Asian influenza. Am J Hyg 68:190–212.
Kafri T, Blomer U, Peterson DA, Gage FH, Verma IM. 1997. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat Genet 17:314–317.
Kawaoka Y, Chambers TM, Sladen WL, Webster RG. 1988. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? J Virol 163:247–250.
Kilbourne ED. 1975. Epidemiology of influenza. In: Kilbourne ED, ed. The Influenza Viruses and Influenza. New York: Academic Press. Pp. 483–538.
Kilbourne ED. 1987. Influenza. New York, NY: Plenum Medical Book Co. Pp. 229–232.
Ko JH, Jin HK, Asano A, Takada A, Ninomiya A, Kida H, Hokiyama H, Ohara M, Tsuzuki M, Nishibori M, Mizutani M, Watanabe T. 2002. Polymorphisms and the differential antiviral activity of the chicken Mx gene. Genome Res 12:595–601.
Ko JH, Takada A, Mitsuhashi T, Agui T, Watanabe T. 2004. Native antiviral specificity of chicken Mx protein depends on amino acid variation at position 631. Anim Genet 35:119–122.
Kodihalli S, Kobasa DL, Webster RG. 2000. Strategies for inducing protection against avian influenza A virus subtypes with DNA vaccines. Vaccine 18:2592–2599.
Krol J, Sobczak K, Wilczynska U, Drath M, Jasinska A, Kaczynska D, Krzyzosiak WJ. 2004. Structural features of microRNA (miRNA) precursors and their relevance to miRNA biogenesis and small interfering RNA/short hairpin RNA design. J Biol Chem 279(40):42230-42239.
Krug RM, Shaw M, Broni B, Shapiro G, Haller O. 1985. Inhibition of influenza viral mRNA synthesis in cells expressing the interferon-induced Mx gene product. J Virol 56:201–206.
Landis H, Simon-Jodicke A, Kloti A, Di Paolo C, Schnorr JJ, Schneider-Schaulies S, Hefti HP, Pavlovic J. 1998. Human MxA protein confers resistance to Semliki Forest virus and inhibits the amplification of a Semliki Forest virus-based replicon in the absence of viral structural proteins. J Virol 72:1516–1522.
Lawson CM, Bennink JR, Restifo NP, Yewdell JW, Murphy BR. 1994. Primary pulmonary cytotoxic T-lymphocytes induced by immunization with a vaccinia virus recombinant expressing influenza A virus nucleoprotein peptide do not protect mice against challenge. J Virol 68:3505–3511.
Lee MT, Klumpp K, Digard P, Tiley L. 2003. Activation of influenza virus RNA polymerase by the 5' and 3' terminal duplex of genomic RNA. Nucleic Acids Res 31(6):1624–1632.
Li WX, Li H, Lu R, Li F, Dus M, Atkinson P, Brydon EW, Johnson KL, Garcia-Sastre A, Ball LA, Palese P, Ding SW. 2004. Interferon antagonist proteins of influenza and vaccinia viruses are suppressors of RNA silencing. Proc Natl Acad Sci USA 101:1350–1355.
Li X, Palese P. 1992. Mutational analysis of the promoter required for influenza virus virion RNA synthesis. J Virol 66:4331–4338.
Liang S, Mozdzanowska K, Palladino G, Gerhard W. 1994. Heterosubtypic immunity to influenza type A virus in mice: Effector mechanisms and their longevity. J Immunol 152:1653–1661.
Liu AN, Mohammed AZ, Rice WR, Fiedeldey DT, Liebermann JS, Whitsett JA, Braciale TJ, Enelow RI. 1999. Perforin-independent CD8+ T-cell mediated cytotoxicity of alveolar epithelial cells is preferentially mediated by tumor necrosis factor-alpha. Relative insensitivity to Fas ligand. Am Respiratory Cell Mol Biol 20:849–858.
Livak KJ, Flood SJ, Marmaro J, Giusti W, Deetz K. 1995. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl 4:357–362.
Lu LV, Askonas BA. 1980. Cross-reactivity for different type A influenza viruses of a cloned T-killer cell line. Nature 288:164–165.
Lu X, Tumpey TM, Morken T, Zaki SR, Cox NJ, Katz JM. 1999. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J Virol 73:5903–5911.
Lukacher AE, Braciale VL, Braciale TJ. 1984. In vivo effector function of influenza virus specific cytotoxic T-lymphocyte clones is highly specific. Exp Med 160:814–826.
Luo G, Danetz S, Krystal M. 1997. Inhibition of influenza viral polymerases by minimal viral RNA decoys. J Gen Virol 78:2329–2333.
Luytjes W, Krystal M, Enami M, Parvin JD, Palese P. 1989. Amplification, expression, and packaging of a foreign gene by influenza virus. Cell 59:1107–1113.
Mazanec MB, Kaeczel CS, Lamm ME, Fletcher D, Nedrud JG. 1992. Intracellular neutralization of virus by immunoglobulin A antibodies. Proc Natl Acad Sci USA 89:6901–6905.
Mazanec MB, Coudret CL, Fletcher DR. 1995. Intracellular neutralization of influenza virus by immunoglobulin A antihemagglutinin monoclonal antibodies. J Virol 69:1339–1343.
Mbawuike IN, Six HR, Cate TR, Couch RB. 1990. Vaccination with inactivated influenza A virus during pregnancy protects neonatal mice against lethal challenge by influenza A viruses representing three subtypes. Virology 64:1370–1374.
Mbawuike IN, Dillon SB, Demuth SG, Jones CS, Cate TR, Couch RB. 1994. Influenza A subtype cross-protection after immunization of outbred mice with a purified chimeric NS1/HA2 influenza virus protein. Vaccine 12:1340–1349.
Mbawuike IN, Pacheco S, Acuna CL, Switzer KC, Zhang Y, Harriman GR. 1999. Mucosal immunity to influenza without IgA: An IgA knockout mouse model. J Immunol 162:2530–2537.
McGrew MJ, Sherman A, Ellard FM, Lillico SG, Gilhooley HJ, Kingsman AJ, Mitrophanous KA, Sang H. 2004. Efficient production of germline transgenic chickens using lentiviral vectors. EMBO Rep 5:728–733.
McMichael AJ, Gotch FM, Noble GR, Beare PAS. 1983a. Cytotoxic T-cell immunity to influenza. N Engl J Med 309:13–17.
McMichael AJ, Gotch FM, Dongworth DW, Clark A, Potter CW. 1983b. Declining T-cell immunity to influenza, 1977–1982. Lancet 2:762–764.
Medema JK, Zoellner YF, Ryan J, and Palache AM. 2004. Modeling pandemic preparedness scenarios: Health economic implications of enhanced pandemic vaccine supply. Virus Res 103:9–15.
Melen K, Ronni T, Broni B, Krug RM, von Bonsdorff CH, Julkunen I. 1992. Interferon-induced Mx proteins form oligomers and contain a putative leucine zipper. J Biol Chem 267:25898–25907.
Meltzer MI, Cox NJ, Fukuda K. 1999. The economic impact of pandemic influenza in the United States: Priorities for intervention. Emerg Infect Dis 5:659–671.
Mibayashi M, Nakad K, Nagata K. 2002. Promoted cell death of cells expressing human MxA by influenza virus infection. Microbiol Immunol 46:29–36.
Morein B, Sundquist B, Hoglund S, Dalsgaard K, Osterhaus A. 1984. Iscom, a novel structure for antigenic presentation of membrane proteins from enveloped viruses. Nature 308:457–460.
Mozdzanowska K, Maiese K, Gerhard W. 2000. Th cell-deficient mice control influenza virus infection more effectively than Th- and B-cell-deficient mice. Evidence for a Th-independent contribution by B-cells to virus clearance. J Immunol 164:2635–2643.
Murphy BR, Coelingh K. 2002. Principles underlying the development and use of live attenuated cold-adapted influenza A and B virus vaccines. Viral Immunology 15:295–323.
Murphy BR, Webster RG. 1990. Orthomyxoviruses. In: Fields BN, Knipe DM et al., eds. Virology. 2nd ed. New York, NY: Raven Press. Pp. 1091–1152.
Murphy BR, Webster RG. 1996. Orthomyxoviruses. In: Fields BN, Knipe DM, Howley PM, eds. Fields Virology. 3rd ed. Philadelphia, PA: Lippincott-Raven. Pp. 1397-1445.
Naldini L, Blomer U, Gage FH, Trono D, Verma IM. 1996. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA 93:11382–11388.
Neirynck S, Deroo T, Saelens X, Vanlandschoot P, Jou WM, Fiers W. 1999. A universal influenza A vaccine based on the extracellular domain of the M2 protein. Nature Med 5:1157–1163.
Neumann G, Hobom G. 1995. Mutational analysis of influenza virus promoter elements in vivo. J Gen Virol 76:1709–1717.
Nguyen HH, Moldoveanu Z, Novak MJ, van Ginkel FW, Ban E, Kiyono H, McGhee JR, Mestecky J. 1999. Heterosubtypic immunity to lethal influenza A virus infection is associated with virus-specific CD8+ cytotoxic T-lymphocyte responses induced in mucosa-associated tissues. Virology 254:50–60.
Nguyen HH, van Ginkel FW, Vu HL, McGhee JR, Mestecky J. 2001. Heterosubtypic immunity to influenza A virus infection requires B-cells but not CD8+ cytotoxic T-lymphocytes. Infect Dis 183:368–376.
Nichol KL, Wuorenma J, von Sternberg T. 1998. Benefits of influenza vaccination for low-, intermediate-, and high-risk senior citizens. Arch Intern Med 80:1769–1776.
Nicholson KG, Tyrrell DA, Harrison P, Potter CW, Jennings R, Clark A, Schild GC, Wood JM, Yetts R, Seagroatt V, Huggins A, Anderson SG. 1979. Clinical studies of monovalent inactivated whole virus and subunit A/USSR/77 (H1N1) vaccine: Serological responses and clinical reactions. J Biol Stand 7(2):123–136.
Nicholson KG, Colegate AE, Podda A, Stephenson I, Wood J, Ypma E, Zambon MC. 2001. Safety and antigenicity of non-adjuvanted and MF59-adjuvanted influenza A/Duck/ Singapore/97 (H5N3) vaccine: A randomised trial of two potential vaccines against H5N1 influenza. Lancet 357:1937–1943.
Obar RA, Collins CA, Hammarback JA, Shpetner HS, Vallee RB. 1990. Molecular cloning of the microtubule-associated mechanochemical enzyme dynamin reveals homology with a new family of GTP-binding proteins. Nature 347:256–261.
Odagiri T, Tashiro M. 1997. Segment-specific noncoding sequences of the influenza virus genome RNA are involved in the specific competition between defective interfering RNA and its progenitor RNA segment at the virion assembly step. J Virol 71:2138–2145.
Offringa DP, Tyson-Medlock V, Ye Z, Levandowski RA. 2000. A comprehensive systematic approach to identification of influenza A virus genotype using RT-PCR and RFLP. J Virol Methods 88:15–24.
Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. 2002. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev 16:948–958.
Parvin JD, Palese P, Honda A, Ishihama A, Krystal M. 1989. Promoter analysis of influenza virus RNA polymerase. J Virol 63:5142–5152.
Patriarca PA, Strikas RA, Gensheimer KF, Cox NJ, Fukuda K, Meltzer MI. 1999. A Pandemic Influenza Planning Guide for State and Local Officials. Draft 2.1. National Vaccine Program Office. Centers for Disease Control and Prevention, Atlanta, GA.
Pavlovic J, Haller O, Staeheli P. 1992. Human and mouse Mx proteins inhibit different steps of the influenza virus multiplication cycle. J Virol 66:2564–2569.
Perdue ML. 2003. Molecular diagnostics in an insecure world. Avian Diseases 47(3 Suppl):1063–1068.
Peters CJ. 2002. Many viruses are potential agents of bioterrorism. ASM News 68:168–173.
Potter CW. 1998. Chronicle of influenza pandemics. In: Nicholson KG, Webster RG, Hay AJ, eds. Textbook of Influenza. London, England: Blackwell Sciences. Pp. 3–18.
Rapp JC, Harvey AJ, Speksnijder GL, Hu W, Ivarie R. 2003. Biologically active human interferon alpha-2b produced in the egg white of transgenic hens. Transgenic Res 12:569–575.
Raulet DH. 1994. MHC class I-deficient mice. 1994. Adv Immunol 55:381–421.
Renegar KB. 1992. Influenza virus infections and immunity: A review of human and animal models. Lab Anim Sci 42:222–232.
Renegar KB, Small PA. 1991. Passive transfer of local immunity to influenza virus infection by 19A antibody. J Immunol 146:1972–1978.
Rhodes GH, Dwarki VJ, Abai AM, Felgner J, Felgner PL, Gromkowski SH, Parker SE. 1993. Injection of expression vectors containing viral genes induces cellular, humoral, and protective immunity. In: Chanock RM, Brown F, Ginsberg HS, Norrby E, eds. Vaccines 93. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. Pp. 137–141.
Rimmelzwaan GF, Baars M, van Amerongen G, van Beek R, Osterhaus AE. 2001. A single dose of an ISCOM influenza vaccine induces long-lasting protective immunity against homologous challenge infection but fails to protect Cynomolgus macaques against distant drift variants of influenza A (H3N2) viruses. Vaccine 20:158–163.
Robertson JS. 1979. 5' and 3' terminal nucleotide sequences of the RNA genome segments of influenza virus. Nucleic Acids Research 6:3745–3757.
Rodgers P, Whitby S, Dando M. 1999. Biological warfare against crops. Sci Am 280:70–75.
Rubinson DA, Dillon CP, Kwiatkowski AV, Sievers C, Yang L, Kopinja J, Rooney DL, Ihrig MM, McManus MT, Gertler FB, Scott ML, Van Parijs L. 2003. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 33:401–406.
Saito T, Lim W, Suzuki T, Suzuki Y, Kida H, Nishimura SI, Tashiro M. 2001. Characterization of a human H9N2 influenza virus isolated in Hong Kong. Vaccine 20:125–133.
Sambhara S, Kurichh A, Miranda R, Tumpey T, Rowe T, Renshaw M, Arpino R, Tamane A, Kandil A, James O, Underdown B, Klein M, Katz J, Burt D. 2001. Heterosubtypic immunity against human influenza A viruses, including recently emerged avian H5 and H9 viruses, induced by FLU-ISCOM vaccine in mice requires both cytotoxic T-lymphocyte and macrophage function. Cell Immunol 211:143–153.
Schnorr JJ, Schneider-Schaulies S, Simon-Jodicke A, Pavlovic J, Horisberger MA, ter Meulen V. 1993. MxA-dependent inhibition of measles virus glycoprotein synthesis in a stably transfected human monocytic cell line. J Virol 67:4760–4768.
Scholtissek C. 1983. Genetic relatedness of influenza viruses (RNA and protein). In: Palese P, Kingsbury DW, eds. Genetics of Influenza Viruses. New York, NY: Springer-Verlag. Pp. 99–126.
Scholtissek C, Burger H, Kistner O, Shortridge KF. 1985. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 147:287–294.
Schulman JL, Kilbourne ED. 1965. Induction of partial specific heterotypic immunity in mice by a single infection with influenza A virus. Bacteriol 89:170–174.
Schwartz-Cornil I, Benureau Y, Greenberg H, Hendrickson BA, Cohen J. 2002. Heterologous protection induced by the inner capsid proteins of rotavirus requires transcytosis of mucosal immunoglobulins. Viral 76:8110–8117.
Scuffham PA, West PA. 2002. Economic evaluation for strategies for the control and management of influenza in Europe. Vaccine 20:2562–2578.
Selin LK, Nahill SR, Welsh RM. 1994. Cross-reactivities in memory cytotoxic T lymphocyte recognition of heterologous viruses. Exp Med 179:1933–1943.
Senne DA. 2003. Avian influenza in the Western Hemisphere including the Pacific Islands and Australia. Avian Diseases 47:798–805.
Seong BL, Brownlee GG. 1992. A new method for reconstituting influenza polymerase and RNA in vitro: A study of the promoter elements for cRNA and vRNA synthesis in vitro and viral rescue in vivo. Virology 186:247–260.
Sims LD, Ellis TM, Liu KK, Dyrting K, Wong H, Peiris M, Guan Y, Shortridge KF. 2003. Avian influenza in Hong Kong 1997–2002. Avian Diseases 47:832–838.
Slepushkin AN. 1959. The effect of a previous attack of Al influenza on susceptibility to A2 virus during the 1957 outbreak. Bull WHO 20:297–301.
Slepushkin VA, Katz JM, Black RA, Gamble WC, Rota PA, Cox NJ. 1995. Protection of mice against influenza A virus challenge by vaccination with barulovirus-expressed M2 protein. Vaccine 13:1399–1402.
Sonoguchi T, Naito H, Hara M, Takeuchi Y, Fukumi H. 1985. Cross-subtype protection in humans during sequential overlapping and/or concurrent epidemics caused by H3N2 and H1N1 influenza viruses. Infect Dis 151:81–88.
Staeheli P, Horisberger MA, Haller O. 1984. Mx-dependent resistance to influenza viruses is induced by mouse interferons alpha and beta but not gamma. Virology 132:456–461.
Staeheli P, Haller O, Boll W, Lindenmann J, Weissmann C. 1986. Mx protein: Constitutive expression in 3T3 cells transformed with cloned Mx cDNA confers selective resistance to influenza virus. Cell 44:147–158.
Steinhoff MC, Fries LF, Karron RA, Clements ML, Murphy BR. 1993. Effect of heterosubtypic immunity on infection with attenuated influenza-A virus vaccines in young children. Clin Microbiol 31:836–838.
Strikas RA, Wallace GS, Myers MG. 2002. Influenza pandemic preparedness action plan for the United States: 2002 update. CID 35:590–596.
Suarez DL, Perdue ML, Cox N, Rowe T, Bender C, Huang J, Swayne DE. 1998. Comparisons of highly virulent H5N1 influenza A viruses isolated from humans and chickens from Hong Kong. J Virol 72:6678–6688.
Subbarao K, Klimov A, Katz J, Regnery H, Lim W, Hall H, Perdue M, Swayne D, Bender C, Huang J, Hemphill M, Rowe T, Shaw M, Xu X, Fukuda K, Cox N. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393–396.
Subbarao K, Chen H, Swayne D, Mingay L, Fodor E, Brownlee G, Xu X, Lu X, Katz J, Cox N, Matsuoka Y. 2003. Evaluation of a genetically modified reassortant H5N1 influenza A virus vaccine candidate generated by plasmid based reverse genetics. Virology 305:192–200.
Takada A, Kuboki N, Okazaki K, Ninomiya A, Tanaka H, Ozaki H, Itamura S, Nishimura H, Enami M, Tashiro M, Shortridge KF, Kida H. 1999. Avirulent Avian influenza virus as a vaccine strain against a potential human pandemic. Virology 73:8303–8307.
Takada A, Matsushita S, Ninomiya A, Kawaoka Y, Kida H. 2003. Intranasal immunization with formalin-inactivated virus vaccine induces a broad spectrum of heterosubtypic immunity against influenza A virus infection in mice. Vaccine 21(23):3212–3218.
Talon J, Horvath CM, Polley R, Basler CF, Muster T, Palese P, Garcia-Sastre A. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol 74:7989–7996.
Taylor SF, Bender BS. 1995. Beta 2-microglobulin-deficient mice demonstrate class II MHC restricted anti-viral CD4+ but not CD8+ CTL against influenza-sensitized autologous splenocytes. Immunol Lett 46:67–73.
Tiley LS, Hagen M, Matthews JT, Krystal M. 1994. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5' ends of the viral RNAs. J Virol 68:5108–5116.
Tompkins SM, Lo CY, Tumpey TM, Epstein SL. 2004. Protection against lethal influenza virus challenge by RNA interference in vivo. Proc Natl Acad Sci USA 101:8682–8686.
Topham DJ, Doherty PC. 1998. Clearance of an influenza A virus by CD4+ T-cells is inefficient in the absence of B-cells. Virology 72:882–885.
Topham DJ, Tripp RA, Doherty PC. 1997. CD8+ T-cells clear influenza virus by perforin or Fas dependent processes. J Immunol 159:5197–5200.
Treanor JJ, Wilkinson BE, Masseoud F, Hu-Primmer J, Battaglia R, O'Brien D, Wolff M, Rabinovich G, Blackwelder W, Katz JM. 2001. Safety and immunogenicity of a recombinant hemagglutinin vaccine for H5 influenza in humans. Vaccine 19:1732–1737.
Tumpey TM, Renshaw M, Clements JD, Katz JM. 2001. Mucosal delivery of inactivated influenza vaccine induces B-cell-dependent heterosubtypic cross-protection against lethal influenza A H5N1 virus infection. J Virol 75(11):5141–5150.
Turan K, Mibayashi M, Sugiyama K, Saito S, Numajiri A, Nagata K. 2004. Nuclear MxA proteins form a complex with influenza virus NP and inhibit the transcription of the engineered influenza virus genome. Nucleic Acids Res 32:643–652.
Ulmer JB, Donnelly JJ, Parker SE, Rhodes GH, Felgner PL, Dwarki VJ, Gromkowski SH, Deck RR, DeWitt CM, Friedman A, Hawe LA, Leander KR, Martinez D, Perry HC, Shiver JW, Montgomery DL, Liu MA. 1993. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science 259:1745–1749.
Ulmer JB, Fu TM, Deck RR, Friedman A, Guan L, DeWitt C, Liu X, Wang S, Liu MA, Donnelly JJ, Caulfield MJ. 1998. Protective CD4+ and CD8+ T-cells against influenza virus induced by vaccination with nucleoprotein DNA. Virology 72:5648–5653.
van Kolfschooten F. 2003. Dutch veterinarian becomes first victim of avian influenza. Lancet 361:1444.
Webster RG. 2002. The importance of animal influenza for human disease. Vaccine 20: S16–S20.
Webster RG, Askonas BA. 1980. Cross-protection and cross-reactive cytotoxic T-cells induced by influenza virus vaccines in mice. Eur J Immunol 10:396–401.
Webster RG, Kawaoka Y, Taylor J, Weinberg R, Paoletti E. 1991. Efficacy of nucleoprotein and haemagglutinin antigens expressed in fowlpox virus as vaccine for influenza in chickens. Vaccine 9:303–308.
Webster RG, Sharp GB, Claas EC. 1995. Interspecies transmission of influenza viruses. Am J Respir Crit Care Med 152:S25–S30.
Wells MA, Albrecht P, Ennis FA. 1981. Recovery from a viral respiratory infection. Influenza pneumonia in normal and T deficient mice. J Immunol 126:1036–1041.
WHO (World Health Organization). 1999. Influenza Pandemic Preparedness Plan. The Role of WHO and Guidelines for National or Regional Planning. Geneva, Switzerland: WHO. [Online]. Available: http://www.who.int/csr/resources/publications/influenza/WHO_CDS_CSR_EDC_99_1/en/ [accesed December 21, 2004].
WHO. 2002. Draft WHO Guidelines on the Use of Vaccines and Antivirals during Influenza Pandemics. Issued 2-4 October 2002. Geneva, Switzerland: WHO. [Online]. Available: http://www.who.int/emc/diseases/flu/whoguidelines.html.
WHO. 2003. Preparing for the Next Influenza Season in a World Altered by SARS. Disease Outbreak #l. Severe Acute Respiratory Syndrome. Update 94 #20. 7-3-2003. [Online]. Available: http://www.who.int/csr/don/2003_07_03/en/ [accessed December 21, 2004].
Wilson TM, Logan-Henfrey L, Weller R, Kellman B. 2000. Agroterrorism, biological crimes and biological warfare targeting animal agriculture. In: Brown C, Bolin C, eds. Emerging Diseases of Animals. Washington, DC: ASM Press. Pp. 23–57.
Wilson TM, Gregg DA, King DJ, Noah DL, Perkins LE, Swayne DE, Inskeep W. 2001. Agroterrorism, biological crimes, and biowarfare targeting animal agriculture. The clinical, pathologic, diagnostic, and epidemiologic features of some important animal diseases. Clin Lab Med 21:549–591.
Wright PF, Webster RG. 2001. Orthomyxoviruses. In: Knipe DM, Howley PM, Griffin DE, eds. Fields Virology. 4th ed. Philadelphia, PA: Lippincott Williams & Wilkins. Pp. 1533–1579.
Xu X, Cox NJ, Bender CA, Regnery HL, Shaw MW. 1996. Genetic variation in neuraminidase genes of influenza A (H3N2) viruses. Virology 224:175–183.
Yetter RA, Lehrer S, Ramphal R, Small PA Jr. 1980. Outcome of influenza infection: Effect of site of initial infection and heterotypic immunity. Infect Immun 29:654–662.
Yewdell JW, Bennink JR, Smith GL, Moss B. 1985. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T-lymphocytes. Proc Natl Acad Sci USA 82:1785–1789.
Zambon M. 1998. Laboratory diagnosis of influenza. In: Nicholson KG, Webster RG, Hay AJ, eds. Textbook of Influenza. London, England: Blackwell Science. Pp. 3–18.
Zhao MQ, Arnir MK, Rice WR, Enelow RI. 2001. Type II pneumocyte-CD8+ T-cell interactions—relationship between target cell cytotoxicity and activation. Am Respir Cell Mol Biol 25:362–369.
Zurcher T, Pavlovic J, Staeheli P. 1992. Mouse Mx2 protein inhibits vesicular stomatitis virus but not influenza virus. Virology 187:796–800.





























































