2
Surveillance Networks
OVERVIEW
As several of the contributors to this chapter note, clinical surveillance of infectious disease is inadequate in much of the developing world due to limited funding for public health infrastructure. Because many impoverished regions are also at high risk for emerging disease threats, alternative methods of surveillance are crucial to global health. The papers collected in this chapter describe a variety of electronic surveillance networks, designed to gather and integrate information on infectious disease from a variety of nontraditional sources (e.g., Internet sites, news outlets, observers with little or no medical training) and to disseminate alerts broadly and rapidly.
The chapter begins with a description of the first infectious disease surveillance network, ProMED-mail. Stephen Morse, one of the network’s founding members, provides a brief history of the free, nonprofit, noncommercial, moderated e-mail list that today serves over 37,000 subscribers in more than 150 countries, as well as anyone with Internet access. Since it began as an experimental system in 1993, ProMED-mail has helped to demonstrate the power of networks and the feasibility of designing effective, low-cost global reporting systems. It has also encouraged the development of additional electronic surveillance networks—such as the Global Public Health Information Network (GPHIN) and HealthMap, described in subsequent contributions to this chapter—and the World Health Organization’s (WHO’s) “network of networks,” the Global Outbreak Alert and Response Network, or GOARN (see Summary and Assessment).
The chapter’s second paper, by presenter Abla Mawadeku and coauthors from GPHIN, offers descriptive comparisons of that network along with ProMED-mail
and the European Commission’s Medical Intelligence System (MedISys), which is available only to European Union member states. GPHIN, a primary source of electronic surveillance for WHO, also serves a host of government institutions, nongovernmental agencies and organizations, academic institutions, and private companies, who pay between 30,000 and 200,000 Canadian dollars per year in subscription fees, depending on the specific services provided.
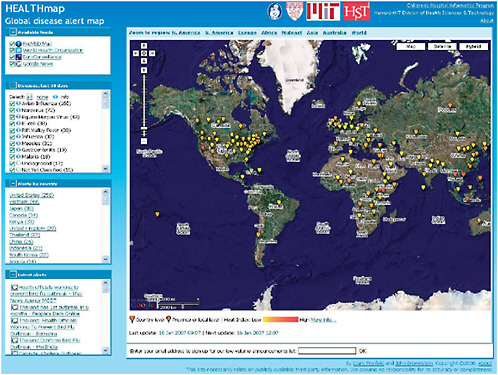
HealthMap is a freely accessible, automated network that collects information from multiple web-based data sources on infectious outbreaks (currently news wires, Really Simple Syndication (RSS) feeds, ProMED mailing lists, and EuroSurveillance and WHO alerts). The network then organizes and displays this information in real time as graphic “maps” featuring geography, time, and infectious disease agent. In their contribution to this chapter, workshop presenter John Brownstein of Harvard Medical School and his colleagues at Children’s Hospital Boston discuss their efforts to evaluate the HealthMap system with reference to four characteristics that have been used to evaluate syndromic surveillance systems: data acquisition; information characterization; signal interpretation; and dissemination. The authors’ preliminary evaluation of HealthMap according to these criteria appears to demonstrate that the aggregation of multiple sources of data—each potentially biased or otherwise flawed—increases the sensitivity and timeliness of alerts while reducing false alarms.
The concluding paper of the chapter describes a different sort of electronic surveillance network: one powered by cell phones, enabling observers in some of the world’s most remote and impoverished communities to report disease outbreaks. The authors are workshop speakers Pamela Johnson of Voxiva, a company that provides information technology to establish surveillance networks in low-resource settings, and David Blazes, of the U.S. Naval Medical Research Center Detachment in Lima, Peru, which used an Internet- and cell phone-based electronic system developed by Voxiva to support disease surveillance by the Peruvian navy along that country’s coast and remote rivers. This experience is presented as a case study in surveillance and evaluated according to the Centers for Disease Control and Prevention (CDC) guidelines for public health surveillance systems. The authors also share lessons gleaned from six years of building surveillance systems, based on cell phones and other cost-effective information technologies, for use in low-resource environments.
Workshop participants raised a series of issues in response to the presentations upon which the papers in this chapter are based. A detailed account of this discussion appears in the Summary and Assessment section, “Considerations for Surveillance Networks.” Discussants were especially concerned about the potentially devastating economic consequences to a country—particularly a developing country—of being labeled (accurately or inaccurately) as harboring a feared infectious disease. In his contribution to Chapter 4, speaker Will Hueston assesses the tradeoff between health and development inherent in the release of surveillance information such as HealthMap’s geographic depictions of outbreak reports.
GLOBAL INFECTIOUS DISEASE SURVEILLANCE AND EARLY WARNING SYSTEMS: PROMED AND PROMED-MAIL
Stephen S. Morse, Ph.D.1
Columbia University
A number of emerging infections have appeared throughout the world in recent years (Morens et al., 2004; IOM, 1992, 2003; Morse, 1995). Or, in the words of Marci Layton (New York City Department of Health and Mental Hygiene), we must learn to expect the unexpected. It is widely agreed that one of the most important measures for both emerging and existing infectious diseases is an effective early warning system, that is to say, global infectious disease surveillance. Here, I will discuss ProMED, the nonprofit international Program for Monitoring Emerging Diseases, and its best known progeny, ProMED-mail (PMM). ProMED itself was founded in 1993 to design and help implement global surveillance systems that could detect both known and emerging infections (Morse et al., 1996).
A Brief History of ProMED and ProMED-Mail
ProMED had its roots in the same Institute of Medicine (IOM) report that led to the development of the Forum on Microbial Threats (IOM, 1992). The Committee that developed the 1992 IOM report was chaired by Joshua Lederberg and the late Robert E. Shope. After the report was released, there was considerable concern about maintaining the momentum. Many of the original Committee members (including me) believed the problem required long-term attention. In addition, for specific reasons the charge to the IOM Committee and consequently the report were limited to the United States. However, there was a clear need to consider these infections as global threats that would require international solutions. In an attempt to fill what many (including this author) saw as the fragmentation of disease surveillance systems and the lack of global capacity, ProMED was begun in 1993 under the auspices of the Federation of American Scientists (FAS).2
Several years earlier, I had been asked by Barbara Hatch Rosenberg, then chairing a working group on biological nonproliferation issues at FAS, to provide technical advice for her working group. After the 1989 National Institutes of Health (NIH) meeting on emerging viruses and the 1992 IOM report, Rosenberg and I discussed the possibility of developing an initiative for global infectious
|
1 |
Mailman School of Public Health. |
|
2 |
An article on the early history and activities of ProMED is available at http://www.fas.org/faspir/pir1293.html, with an update at http://fas.org/promed/announce.htm. Additional materials are available at http://fas.org/promed/. |
disease surveillance, with start-up resources from FAS. Dorothy Preslar served as the project staff at FAS.
The group held a small initial organizational meeting in February 1993 at The Rockefeller University in New York. In addition to Rosenberg, and myself as Chair, among those present at that meeting were Ruth Berkelman (then at the Centers for Disease Control and Prevention, or CDC), Scott Halstead (then at the Rockefeller Foundation), D. A. Henderson (then at Johns Hopkins and the U.S. Department of Health and Human Services), James M. Hughes (then at CDC), John LaMontagne (then at NIH’s National Institute of Allergy and Infectious Diseases, or NIAID), and Shope (then at Yale). At that time, it was decided that a conference would be held in Geneva in fall 1993, that the group’s purview should include animal and plant diseases in addition to human disease (a view especially advocated by Berkelman), and that the group should be named ProMED (Shope suggested the name).
The next activity was a conference, cosponsored by FAS and the World Health Organization (WHO) and held on September 11 and 12, 1993, at WHO headquarters in Geneva. Part of the challenge at that time was that the then-Director General of WHO did not believe that surveillance for infectious diseases was part of the organization’s core responsibilities. Unfortunately, many clinicians and most of the lay public naïvely believed otherwise, and thought that WHO was already doing it.
The September 1993 ProMED meeting, co-chaired by Francis Nkrumah of Ghana and myself, was held in the WHO Executive Board Room, and included as speakers a number of people who had been influential in WHO affairs, including Jan Kostrzewski, a former chair of the WHO Executive Board, Henderson, and a number of members of the World Health Assembly. At that event, 60 prominent scientists and public health officials working on human, animal, and plant health from all parts of the world met, unanimously endorsed the concept of global surveillance, and formed ongoing working groups to assess present capabilities and develop and implement plans for a suitable global program that could address both known and emerging infections. We also invited John P. (Jack) Woodall (then at WHO) onto the Steering Committee, and James LeDuc (then at WHO, seconded from CDC) agreed to serve as a special consultant.3
One would think it should be fairly simple to strengthen and network regional centers of excellence to augment official systems and develop mutual cooperation, whether through WHO (preferably) or through regional intergovernmental organizations. On the other hand, if diseases can emerge anywhere, how can one get early warning from literally everywhere? The latter seemed the harder task, so we decided to try tackling what everyone considered the easier one first. At meetings in Geneva and elsewhere, we recommended developing a coordinated
|
3 |
The list of the early Steering Committee members can be found at http://fas.org/promed/about/steering.html. |
system of regional centers and a minimum set of capabilities to identify and respond to unusual disease outbreaks. A plan was subsequently published (Morse et al., 1996), in part elaborating on the system Henderson had proposed at the 1989 NIH/NIAID meeting on emerging viruses (Henderson, 1993). The strategy developed was vigilance for unusual clinical presentations of special concern (e.g., encephalitis or acute respiratory distress with fever in adults); a minimum set of microbiological capabilities at each site to identify common diseases; and a system to refer unidentifiable samples to successively more sophisticated reference laboratories, through the network, for possible identification. The plan also included epidemiologic capacity, which could be provided rapidly through the network if needed (Morse et al., 1996).
The effort continued with meetings at other places. At a Steering Committee meeting in June 1994 at Airlie House in Virginia, we realized that our members from all over the world had no reliable means to communicate with one another. Nkrumah of Ghana, for example, had a Telex, which in any major American city usually required a trip downtown to a special office building to send, but no fax machine. In Russia, they had fax machines but no fax paper because of a lack of money. We decided to try to put everyone on a common communications system. Charles Clements, then at a nonprofit organization called SatelLife, which specialized in inexpensive e-mail connections for remote and underserved areas through satellite radio links, had been invited to the meeting. I appointed Woodall as head of a new Communications Task Force. By the end of the meeting a plan had been developed to connect everyone by e-mail. SatelLife provided connectivity for places without e-mail connections, for example (at that time) in Africa, China, and Russia. The rest of us learned how to use the existing e-mail systems at our institutions (quite an ordeal in those days). Thus ProMED-mail was born. Although only about 10 years ago, it was another era technologically.
As the system developed and people started using e-mail for communications, we realized it could also be used as an international outbreak reporting system. (So much for deferring those “more challenging” goals, such as how to get reports from everywhere.) Woodall and I served as the initial moderators (or “editors”), a time-consuming task. Woodall deserves tremendous credit for his dedication and enormous contributions to the subsequent development of the system. Since 1995, the system has been available on the Web,4 as well as by e-mail subscription. The partnership between ProMED and SatelLife continued fruitfully until 1999, when the ProMED reporting network was transferred to the International Society for Infectious Diseases (ISID), headquartered at Harvard’s Channing Laboratory in Boston. The communications network was renamed ProMED-mail, to distinguish it from other ProMED activities then underway.
ProMED-Mail: A Prototype Infectious Disease Reporting System
Many people think of PMM as synonymous with ProMED, as it has taken on a robust life of its own. PMM was designed as an open reporting and discussion system. It is a nonprofit, noncommercial e-mail list that now has some 40,000 subscribers, with over 165 countries represented. Not all of them, of course, send in reports because the editors would be overwhelmed, but many subscribers do read the e-mails on a regular basis. Although numbers vary, incoming e-mails (roughly 100 a day) generate an average of 7 to 10 reports every day.
The e-mail listserv is moderated, which means that messages coming in are first read by people with scientific or medical expertise. Originally this was Woodall and at times me until I left for government service in 1996. As the list grew, a number of other moderators were recruited in various specialty areas, and the system is fortunate to have a number of distinguished experts as moderators.
In principle, subscribers send in reports and information. Rapporteurs take additional responsibility to report regularly in their own geographic or special interest areas. Rapporteurs report from Russia, China, and a number of other places as well as within the United States. When someone sends in a report from somewhere (one of the earliest reports of Ebola in Kikwit, Zaire, now the Democratic Republic of Congo, came from a medical missionary who had a radio e-mail link), the report is assigned by the editor-in-chief or someone acting in that capacity, to the appropriate moderators for editing and, if appropriate, posting to the list. The moderator reads the report for scientific plausibility. If the report looks credible, the moderator edits and formats as needed, probably adds comments to put the item in context, and send it out as a posting to the list. All subscribers are free to comment or add information after reading the posting.
In addition to the full list, which includes outbreak reports and discussions on human, veterinary, and plant diseases, there are several sublists for those who want only certain parts of this information. It is possible to subscribe to the animal and plant disease lists separately. The human disease list includes both human and animal disease. This causes occasional complaints from physicians, but we have believed strongly from the beginning that it is essential to improve the connections between animal and human health. Justifying this is the fact that many emerging infections are zoonotic. Those who are interested in getting only the breaking news, without the ensuing discussion, can subscribe to the Emerging Disease Reports (EDR) sublist. I get EDR on my BlackBerry wireless device.
In recent years sub-lists have been developed in Portuguese, Spanish, and Russian, and there is interest in developing other foreign language lists as well. Some of the regional reports of wide interest are translated into English.
The PMM architecture is simple. Technically, the e-mails are 7-bit ASCII text, the most basic format. When the system was started in August 1994, people in developing countries had very limited bandwidth. It is amazing how much
this has changed in the past decade, with broadband Internet cafes now even in remote areas.
The editors also search the Web and press reports, an increasingly important source of information. This strategy was originally adopted by GPHIN (the Canadian government’s Global Public Health Intelligence Network), which is described in another chapter. GPHIN was started in 1999 and is based largely on news sources from the Web. Unfortunately such material was not available when PMM was started. Since then, the explosive growth of the Web and of improved methods for searching have made such strategies very effective.
Perhaps one of the most important value-added features of PMM is the distinguished and hard-working team of moderators or editors (for this chapter, I am using these two terms interchangeably). Although they are essentially volunteers, all are subject-matter experts. The moderators also have their own e-mail lists and personal networks for follow-up, which demonstrates the power of networking. Larry Madoff is the current editor-in-chief of PMM, while Woodall (now associate editor) remains as active and involved as ever. He has had a critical role in developing PMM into what it is today. Eduardo Gotuzzo, in addition to being a member of the IOM Forum, is Chair of the PMM Policy Committee.
All this is probably obvious to anyone who has read PMM. Anyone can contribute; data come from clinicians (those proverbial astute clinicians in the field all over the world), public health officials and epidemiologists, lab scientists, or medical missionaries, but also journalists and interested laypeople.
There was a concern initially that the method of obtaining data would give rise to many rumors that health authorities would then have to verify, expending valuable resources. This has not turned out to be a major problem. Of course, sometimes information is incorrect, but in general the reliability turns out to be more than 95 percent, according to figures that Madoff tabulated. However, PMM has developed several mechanisms to deal with the possibility of erroneous reports. One is personal follow-up by moderators. The moderators, experts in their fields and generally well connected, can use their own personal networks to try to get more information to include. Second, an uncertain report could also be posted as a request for information (RFI), an inquiry which is simply a way of asking people if they have more information they can contribute. Others on the network may also spontaneously add to or correct a posting if they have additional facts.
Subsequently, WHO, in response to information from PMM and GPHIN, developed a very effective mechanism of its own, called the Outbreak Verification List. WHO sends this list out regularly to a limited group of public health officials and scientists to try to follow up on various outbreak reports. It is a sign of WHO’s increasing capacity and interest that the reports increasingly are coming from WHO’s own country and regional representatives. WHO has developed its own network of networks, the Global Outbreak Alert and Response Network (GOARN), which includes a number of formal and informal sources. It should
be noted that the situation at WHO has greatly improved in the last few years, thanks to the concerted efforts of a number of people, including James LeDuc in the early days, and notably David Heymann more recently.
One particularly interesting aspect of a system like PMM is that it can be used to compare reports from a number of places. In addition to outbreak reporting, it provides the ability for people to recognize that what they are observing may be happening elsewhere, too. An initial report may encourage others to contribute local information that may help to estimate the extent and numbers of an infectious disease outbreak, and to monitor spread. One example was a 1995 outbreak of meningococcal meningitis occurring simultaneously in several states and in the United Kingdom. The outbreak became evident when the reports from various places appeared on PMM.
PMM has been available on the Web since the Ebola outbreak of 1995 in Kikwit, when it partnered with and later incorporated an independent effort called “Outbreak.” As the Web itself grows, the website has had an increasing presence. If one prefers not to receive e-mail alerts, it is a simple matter just to search the website and read any of the reports. The Web archives include some of the earliest reports, such as the first reports of Ebola in Kikwit. Among other PMM “firsts” was Venezuelan equine encephalitis, coincidentally in Venezuela. It was originally denied by the government; when it was verified it led to the resignation of the health minister. West Nile virus in 1999 was another event PMM extensively covered. During this period, Ian Lipkin generously wrote in to offer reagents for people internationally. Other firsts include reports of H5N1 influenza in Indonesia in November 2003 and fatalities in China in 2005 attributed to Streptococcus suis.
The first report of severe acute respiratory syndrome (SARS) that appeared on PMM was a rumor about an unusual outbreak in south China with unexplained deaths. Steve Cunnion picked this up, and information was posted on February 10, 2003. Shortly after that, China officially reported the disease, and WHO was able to release information officially. By that time, China reported 305 cases. SARS had actually been infecting people for at least several months (IOM, 2004). SARS then spread to Toronto, where it was originally called “atypical community-acquired pneumonia” and was reported on PMM.
Madoff has tabulated the PMM disease reports over the past 10 years. Dengue, which is quite common, is one constant, as are a number of others. Many are known conditions, but at least 209 are not. Some will eventually be added to the known category. There have also been reports of CDC Category A agents, normally more closely associated with bioterrorism or biowarfare. However, anthrax exists naturally throughout the world in livestock. In developing countries, there may be thousands of cases of gastrointestinal anthrax from contaminated meat. More than 200 cases of anthrax in livestock were reported on PMM before the intentional anthrax attacks of fall 2001. Botulism and tularemia are also natu-
rally occurring diseases, which reminds us that many of the classic bioweapons, including the Category A agents, are zoonotic agents.
PMM was developed as a prototype, and continues to evolve. There have been increasing efforts since then. GPHIN and WHO’s GOARN have already been mentioned. A later paper, by Pamela Johnson, will discuss Voxiva, which uses the power of networks with another technological base, the cell phone network.
PMM has also elicited some kind comments. Henderson referred to CNN and PMM as the major sources of information for infectious diseases. Steven C. Joseph (formerly New York City Health Commissioner, Dean of the Minnesota School of Public Health, and Assistant Secretary of Defense for Health Affairs) referred to PMM as “the CNN of infectious diseases” (Personal communication, S. C. Joseph, June 1995). Perhaps the most intriguing characterization comes from Steven Johnson, in his book The Ghost Map, about cholera in Victorian London. A sentence in the book caught me by surprise as I was leafing through it:
The popular ProMED-mail e-mail list offers a daily update on all the known disease outbreaks flaring up around the world, which surely makes it the most terrifying news source known to man (Johnson, 2006).
For an infectious disease surveillance system, that seems high praise indeed.
Since PMM was started as an experimental system more than a decade ago, it has helped to demonstrate the power of networks and the feasibility of designing widely distributed, low-cost reporting systems, and it has encouraged the development of additional systems using additional technologies. All these efforts help to begin building the heavily networked surveillance systems that will be needed to deal with threats in an increasingly globalized and unpredictable world.
Acknowledgments
Sincere thanks to Jack Woodall and Larry Madoff for their hard work and helpful discussions, and to all the editors/moderators and funders of ProMED-mail for their dedication and good work.5 Stephen S. Morse is supported by CDC cooperative agreements A1010-21/21, U90/CCU224241 (Centers for Public Health Preparedness), and U01/CI000442, the Arts & Letters Foundation, and by NIH/NIAID cooperative agreement 5U54AI057158 (Northeast Biodefense Center RCE).
|
5 |
A list of current PMM personnel is at http://www.promedmail.org; click on “Who’s Who.” |
GLOBAL PUBLIC HEALTH SURVEILLANCE: THE ROLE OF NONTRADITIONAL SURVEILLANCE TOOLS
Abla Mawudeku, M.P.H.6
Public Health Agency of Canada
M. Ruben, M.D., Ph.D.7
Public Health Agency of Canada
R. Lemay, B.Sc., M.B.A.8
Public Health Agency of Canada
Introduction
In a world deeply interconnected by traveling and trade, the spread of infectious agents is inevitable. Regions once isolated are now integrated into the global community and have the risk of being exposed to infectious agents that they previously were unexposed to, as well as sources of old and new agents, and even new pandemics. Therefore, there is global concern about surveillance and control of diseases (particularly infectious diseases) around the globe.
Any global surveillance system has to overcome several challenges; basically, it requires a good system for communications to and from the field to get timely collection, analysis, and dissemination of data, and to be able to force political decisions and allocation of resources. However, susceptibility to infectious diseases and increased risks of infection are usually associated with poverty, and poverty is more frequent in those countries where epidemiological and laboratory surveillance is defective or nonexistent (Heymann and Rodier, 2001). In addition, while several countries, particularly in the Western world, have already national surveillance systems to monitor for potential public health threats, in many circumstances these systems are inadequate, fairly erratic, or too disease specific to identify new diseases early (Butler, 2006). Also, countries have been reluctant to report outbreaks due to the perception of a negative impact of such news on the country’s economy (trade and tourism). Public alarm, sometimes fueled by the press, has resulted in many occasions in important losses for the countries, which then try to hide or delay the recognition of the presence of human or animal diseases (Cash and Narasimhan, 2000). Nevertheless, the electronic era, in which press reports and the Internet keep societies informed and interconnected, have begun to break down all attempts of “secrecy.”
Currently there is no comprehensive global public health surveillance system. The World Health Organization (WHO) is the only organization that has the mandate to monitor and respond to global public health threats, as established
in the International Health Regulations (IHRs). WHO not only uses information gathered from traditional surveillance systems but also uses information from nontraditional surveillance systems to leverage in order to capture a more comprehensive outlook of the situation about potential public health threats occurring worldwide. The use of nontraditional surveillance systems has contributed to the improvement of epidemic intelligence used for the early detection of potential public health threats. This has enabled WHO and other public health organizations such as the European Center for Disease Control (ECDC) to better assess, investigate, and respond to events of concern (Figure 2-1).
A revised version of these regulations, IHR 2005, will be implemented in June 2007. These new IHRs will strengthen WHO’s authority in surveillance and response because they include more demanding surveillance and response obligations and apply human rights principles to public health interventions (Baker and Fidler, 2006). The new regulations require that member countries report to
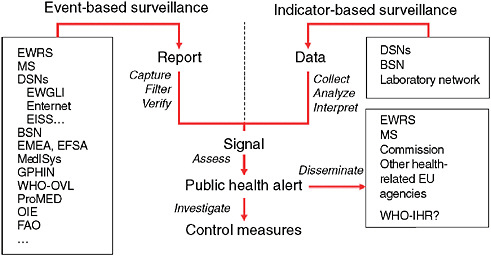
FIGURE 2-1 Epidemic intelligence framework.
EWRS = Early Warning Response System; MS = messaging system; DSN = disease surveillance network; EWGLI = European Working Group for Legionella Infections; EISS = European Influenza Surveillance Scheme; BSN = Basic Surveillance Network; EMEA = European Agency for the Evaluation of Medicinal Products; EFSA = European Food and Safety Authority; WHO-OVL = Outbreak Verification List; OIE = Office International des Epizooties (World Organization for Animal Health); FAO = Food and Agriculture Organization; EU = European Union; and Enter-net is an established and thriving EU-wide network for the laboratory-based surveillance of human Salmonella and Verocytotoxin-producing Escherichia coli (VTEC) infections.
SOURCE: Based on Kaiser et al. (2006).(2006).
WHO “all events which may constitute a public health emergency of international concern” (i.e., unexpected or unusual public health events that might include communicable and noncommunicable disease events, whether natural, accidental, or intentionally created). IHR 2005 also requires from member countries (if practicable) to report to WHO all public health risks identified outside their territories that might cause international disease spread (Baker and Fidler, 2006). They also give WHO more autonomy from the governments of member countries; WHO can now use nontraditional surveillance information (i.e., data from the news media) and ask the countries about “rumors” of circulating infectious agents.
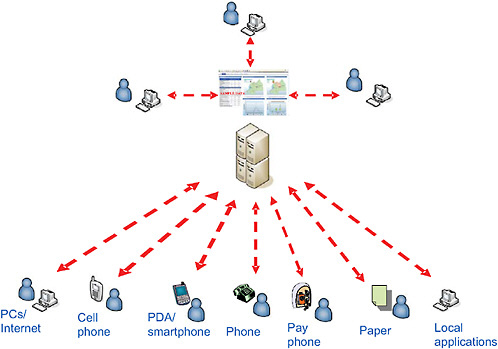
Several innovative nontraditional surveillance systems leverage the advancements in modern Internet and information technologies to efficiently and rapidly gather information about events of public health concern. The Global Public Health Intelligence Network (GPHIN), the Program for Monitoring Emerging Diseases (ProMED), and Medical Intelligence System (MedISys) are examples of such systems that are commonly used by the public health community. All these surveillance systems disseminate relevant reports to the public health community in a timely manner.
Global Public Health Intelligence Network
GPHIN is an early warning system that takes advantage of existing information technology to continuously scrutinize news media sources through news aggregators who have contracts with newspapers around the world, as well as with health and science websites. The multilingual system gathers information by monitoring global media on a 24/7 basis and in nine languages, including Arabic, Chinese (simplified and traditional), English, Farsi, French, Russian, and Spanish. More recently, Portuguese has been added. In addition, and with the help of automated translation software, non-English articles are translated into English, and English articles translated into French, Portuguese, Spanish, Russian, Chinese (simplified and traditional), Farsi, and Arabic. The translations give the essence of the news report.
The system, which has automated and manual components, searches for information on disease outbreaks and other emerging and reemerging public health threats (e.g., contaminated food and water, bioterrorism, chemical or radiological threats, natural disasters) and then generates timely alerts (Figure 2-2). The automated process helps to organize and prioritize the relevant news media reports that are reviewed and analyzed by a team of analysts who are multilingual and multidisciplinary (Figure 2-2). The analysts work in shifts and provide analytical coverage on a 24/7 basis. The analysts have the responsibility of identifying events that may have serious public health consequences, and of flagging them as alerts following preestablished criteria. The analysts also review, periodically, the items kept in the database as irrelevant, to ensure that none of these items represents a potential alert. In addition, the analysts are responsible
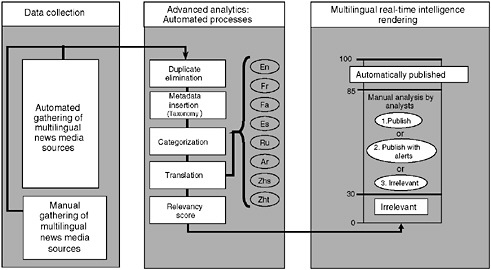
FIGURE 2-2 Global Public Health Intelligence Network (GPHIN) flow of information.
SOURCE: Public Health Agency of Canada.
for identifying trends or relationship of events, checking clarity of machine translations, and updating search syntaxes and keywords used to monitor and gather relevant news media reports. The users are also able to interact with the analysts to request assistance or to provide feedback.
GPHIN is currently one of the primary sources of information for WHO. Other GPHIN users include government institutions, nongovernmental agencies and organizations, as well as academic institutions and private companies that conduct public health surveillance worldwide. Users have access to GPHIN through a password protected website and also receive e-mail alerts (Mykhalovskiy and Weir, 2006).
ProMED-Mail
ProMED9 offers a free public website and an e-mail list that has subscribers from around the world (currently more than 37,000 subscribers from over 150 countries). Its mission is “to provide early warning, 7 days a week year around, of outbreaks of emerging infectious diseases and episodes of acute toxicity, and the spread of antibiotic and disease vector resistance, worldwide, free of charge by e-mail” (Woodall, 2001; Woodall and Calisher, 2001). The system distributes information about outbreaks often early on, before it is confirmed by WHO;
|
9 |
ProMED-mail, International Society for Infectious Diseases, http://www.isid.org. |
therefore, it complements the global surveillance done by WHO and countries (Woodall, 2001).
It allows communications by e-mail all over the world, and includes sublists of reports in Spanish, Portuguese, and Russian, with some of the most interesting local reports translated into English. ProMED publishes media reports, personal reports, and summaries; it presently covers not only human diseases, but also animal and plant diseases, and it is also available on the Web (Woodall, 2001). Most data published by ProMED comes from individuals (clinicians, public health officers, epidemiologists, laboratory scientists, and lay individuals) or from academic or official organizations worldwide.
ProMED has several moderators who cover their own geographic areas (e.g., Russia, China) and search the Web and press reports for relevant news. These moderators are subject-matter experts and provide their expertise as volunteers (Mykhalovskiy and Weir, 2006); they also have their own e-mail lists and personal networks to follow up the reports, and they frequently add their comments and their knowledge to the news.
MedISys
MedISys is a near real-time news alert automated system managed by the Directorate General Health and Consumer Affairs of the European Commission.10 MedISys covers emerging and reemerging public health issues related to communicable diseases and bioterrorism. It monitors on a 24/7 basis approximately 800 Web sources (news and medical sites) daily in 25 languages, including the languages of European Union (EU) member states, Arabic, and Chinese. Access to MedISys is limited to EU member states.
Conclusions
The continuous proliferation of emerging and reemerging pathogens able to infect humans, domestic animals, plants and wildlife seems to have increased in the past years, helped by the increased and faster movement of people and goods. This has generated international concern and increased efforts to improve the early warning capacity to detect potential public health threats worldwide in order to control and prevent the spread of diseases (Heymann and Rodier, 2001; Formenty et al., 2006).
Today’s advancements in communication technology (e.g., blogs, wikkies), and information technology are used liberally by the news media and the public; this makes possible the rapid dissemination of worldwide news about events of public health concern. Such proliferation of information has made it challenging for the public health community, with limited resources, to be aware of and
analyze all the data available in an efficient and effective manner. Current early warning tools, such as GPHIN, MedISys, and ProMED also find it challenging to keep abreast of all the sources of information available. Therefore, the most feasible and cost-effective solution would be to establish a network of nontraditional early warning surveillance systems in order to leverage the expertise provided by each system. GPHIN, MedISys, and ProMED, which are complementary, could then strengthen the mutual abilities of monitoring, gathering, analyzing, and disseminating information about events of public health concern.
In such a collaboration, ProMED’s team of experts would provide reports of relevant events; GPHIN’s would add a team of multilingual, multidisciplinary analysts plus its technical capacity to process high volumes of disparate multilingual data; and MedISys would add its capacity to monitor the Internet for news in more than 20 languages, improving the gathering of information about potential public health threats in remote areas. In addition, this collaboration would make possible the dissemination of synthesized information (from the numerous news sources) about relevant events, highlighting major points and strengthening epidemic intelligence. Furthermore, visualization features, like the ones provided by the Geographic Information System (GIS), could also facilitate the epidemiological analysis of public health threats.
For such a comprehensive and ambitious network to be effective, and to reach the entire planet, it would also need the support of the public health community and wildlife, animal, and agricultural experts, when possible (Jebara, 2004; Butler, 2006). It also would need technologic and economic support from the private sector. It is expected that a network like this could adapt to the needs of the different customers, and provide support to all countries, worldwide, to strengthen their surveillance systems and be able to accomplish the mandate of the IHR guidelines. These guidelines anticipate that each member state should assess its capability to strengthen and maintain core surveillance capacities by 2009 and develop a plan to accomplish this (Hardiman, 2003; Baker and Fidler, 2006).
HEALTHMAP: INTERNET-BASED EMERGING INFECTIOUS DISEASE INTELLIGENCE
John S. Brownstein, Ph.D.11
Harvard Medical School
Clark C. Freifeld, B.S.12
Children’s Hospital Boston
Ben Y. Reis, Ph.D.13
Children’s Hospital Boston
Kenneth D. Mandl, M.D., M.P.H.14
Children’s Hospital Boston
Although many developed countries are strengthening their traditional clinically based surveillance capacities, the required health information infrastructure is lacking in parts of the world that may be most vulnerable to emerging health threats. At the same time, an enormous amount of information providing situational awareness about infectious diseases is found in web-accessible information sources, such as Internet-based discussion sites, disease reporting networks, news outlets, and blogs. These data also exemplify unprecedented potential for increasing public awareness on public health issues prior to their widespread recognition. Despite the growing use of these unstructured information sources for monitoring emerging infectious diseases, there has been little, if any, formal evaluation of their utility, accuracy, coverage, or timeliness. Building on established evaluation approaches for public health surveillance systems, we present a surveillance framework that defines important challenges and critical research questions that define a research agenda. The framework is informed by evaluation of the performance of HealthMap, a freely accessible, automated system for real-time monitoring of online information about emerging diseases. This chapter highlights the value of a robust research agenda, continued organic evolution of existing and new technologies, and scrutiny through a rigorous evaluation framework to help ensure that the global public health enterprise maximally leverages
|
11 |
Instructor, Department of Pediatrics, Harvard Medical School, Children’s Hospital Boston, 1 Autumn St, Room 541, Boston, MA 02215. Phone: 617-355-6998. Fax: 617-730-0267. E-mail: john_brownstein@harvard.edu. |
|
12 |
Research Software Developer, Children’s Hospital Informatics Program at the Harvard–MIT Division of Health Sciences and Technology, Children’s Hospital Boston, 300 Longwood Ave., Boston, MA 02115. |
|
13 |
Instructor, Children’s Hospital Informatics Program at the Harvard–MIT Division of Health Sciences and Technology, Children’s Hospital Boston, 300 Longwood Ave., Boston, MA 02115. |
|
14 |
Associate Professor, Children’s Hospital Informatics Program at the Harvard–MIT Division of Health Sciences and Technology, Children’s Hospital Boston, 300 Longwood Ave., Boston, MA 02115. |
new electronic sources for surveillance, communication, decision making, and intervention.
Introduction
Real-time public health surveillance represents a critical tool for controlling infectious diseases, an effort that requires a timely and global approach addressing the complex and dynamic interactions among infectious agents, animals, and the environment (Heymann and Rodier, 1998, 2001; Woodall, 2001). Although many developed countries are strengthening their traditional clinically based surveillance capacities, the required health information infrastructure is lacking in parts of the world that may be most vulnerable to emerging health threats (Butler, 2006). The existing network of traditional surveillance efforts by health ministries, institutes of public health, multinational agencies, and laboratory and institutional networks has gaps in geographic coverage and often suffers from poor information flow across national borders.
At the same time, an enormous amount of information providing situational awareness about infectious diseases is found in web-accessible information sources, such as Internet-based discussion sites, disease reporting networks, news outlets, and blogs (Heymann and Rodier, 2001; Grein et al., 2000; M’Ikanatha et al., 2006). Even web-based clickstream and keyword searching aggregated across Internet users can provide important insights (Eysenbach, 2006). These resources provide valuable and highly local information about disease outbreaks and related events, even from areas relatively invisible to daily global public health efforts (Woodall, 1997). In fact, the majority of outbreak verifications currently performed by the World Health Organization’s (WHO’s) Global Outbreak Alert and Response Network (GOARN) initially begin as reports from informal electronic data sources such as mailing lists and local news media (Heymann and Rodier, 2001; Grein et al., 2000).
While these web-based data sources can facilitate early detection of outbreaks, they may also support increasing awareness of public health issues prior to their formal recognition. Through low-cost and real-time Internet data mining combined with open-source and user-friendly technologies, participation in global disease surveillance is no longer limited to the public health community (Keystone et al., 2001; Petersen, 2005). Furthermore, the availability of web-based media across national borders greatly ameliorates the potentially suppressive effects of political influence on the spread of information.
The HealthMap Project
Though valuable, electronic sources of emerging infectious disease news are not well organized or integrated. We sought to develop HealthMap, a freely accessible, automated approach to organizing data about infectious outbreaks accord-
ing to geography, time, and infectious disease agent (Figure 2-3) (Holden, 2006; Larkin, 2007; Captain, 2006). HealthMap is a multistream real-time surveillance system that aggregates multiple Web-based data sources (currently news wires, Really Simple Syndication (RSS) feeds, ProMED mailing lists, and EuroSurveillance and WHO alerts). Information is acquired automatically through screen scraping, natural language interpretation, text mining, and parsing to obtain disease name and geocode the location of the outbreak. HealthMap also addresses the computational challenges of integrating multiple sources of unstructured online information in order to generate robust meta-alerts of disease outbreaks. Through this approach, we achieve a unified and comprehensive view of current global infectious disease outbreaks in space and time.
System Challenges
Despite the success of Internet-based surveillance systems such as HealthMap, important technological and methodological challenges remain. Four principal development and deployment issues are as follows:
-
Value. Though there is an abundance of disparate electronic resources, none is comprehensive. Each has gaps in coverage of certain geographic areas, population sectors, medical expertise, and availability.
-
Standards. No universal standards exist for capturing, processing, reporting, interpreting, or sharing structured data. Such standards would greatly facilitate the communication and use of information by computationally based systems.
-
Performance. Metrics for systematic evaluation of these data sources and the performance of these systems are still needed. Though there has been some description of individual data sources (M’Ikanatha et al., 2006; Cowen et al., 2006), there is still limited understanding of their value for spatial and temporal detection and monitoring of disease outbreaks.
-
Accessibility. Important issues require attention to system ownership, target audience, restrictions, cost, and sustainability.
Surveillance Framework
A good starting point for design of a surveillance framework is the one currently used for the syndromic surveillance systems that have evolved over the past eight years (Mandl et al., 2004a; Buehler et al., 2004; CDC, 2000). The anthrax attacks of 2001 gave rise to large-scale surveillance efforts directed at early detection of an outbreak, prior to confirmed diagnosis (Perkins et al., 2002). These novel surveillance systems also use data that are not diagnostic of a disease, but that might indicate the early stages of an outbreak, often earlier than might otherwise be possible with traditional public health methods. The ideal syndromic surveillance system has the following traits: it acquires data automati-
cally; collects ongoing data in real time or near real time; electronically stores and transmits data to an analytic module; has sufficient demographic, geographic, and temporal coverage to support anomaly detection; captures data in standard formats across data sources; protects private information and patient confidentiality; and scans for outbreaks, correctly distinguishing an abnormal pattern from a normal or expected one (Mandl et al., 2004a). While Internet-based surveillance represents a paradigm shift from indicator-based to event-based sources of information, the existing framework is designed to support the evaluation of all public health surveillance systems. The standard set of evaluation metrics used to interpret data quality and signal detection should apply across both traditional and Internet-based surveillance approaches (Mandl et al., 2004a, b; Buehler et al., 2004; Wagner et al., 2001). Both Internet-based surveillance and traditional syndromic surveillance require four stages: (1) data acquisition, (2) information characterization, (3) signal interpretation, and (4) dissemination and alerting (Figure 2-4).
Here we present a summary of initial evaluation efforts based on this surveillance framework. To help inform our evaluation, we analyzed the HealthMap alert data stream, over a 20-week period (October 1, 2006, through February 17, 2007),
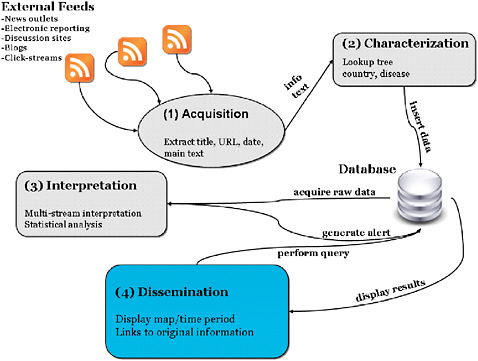
FIGURE 2-4 Framework for Internet-based surveillance.
SOURCE: Brownstein (2006).
applying standard evaluation metrics (volume, geography covered, diseases captured, timeliness, sensitivity, and specificity). Over this period, HealthMap found 3,194 news reports of infectious disease outbreaks (a mean of 22.8 per day, 95 percent confidence interval, 20.6–25.0).
Data Acquisition
Data can be acquired by search either of the open Web or of specific restricted or open websites. The choice of data sources has critical implications for early outbreak detection and disease monitoring across all metrics. Here data sources are evaluated across three dimensions: quality, cost, and availability.
Data Quality
The ideal information sources would be sensitive to even the smallest aberrations. However, as in all surveillance activities, there is an inherent tradeoff between the timeliness and specificity of a system. For example, local news sources may report on strange incidents involving a few cases that would not be picked up at the national level. However, local news reports may be less reliable, reporting stories without adequate confirmation. Information is not always validated, and the credibility of the sources is not always vetted. Thus, without proper filtering, these local news sources may be responsible for substantial noise in the system and increasing the overall false alarm rate. Furthermore, other biases may be introduced for political reasons, resulting either in disinformation (false positives) or censorship (false negatives). In the case of ProMED, its hierarchical curation structure helps minimize false positives. However, while expert review does increase specificity, the required manual processing delays alert reporting.
Data Cost
Internet-based surveillance data have been limited largely to automated mining of information from news aggregators. An important question is whether paid subscription sites provide more value than freely available information. For example, news aggregators such as LexisNexis®, Factiva®, and Magenta News® may all be useful sources of information, especially for local news in a substantial number of languages. However, free online news aggregators, such as Google News and Yahoo News, potentially integrating up to 10,000 sources, may have almost equal value.
The use of free data means that these systems may be provided at minimal cost to the public and to countries that lack the resources to pay high subscription rates. For paid data, cost-effectiveness of various data sources becomes an important issue. Data for HealthMap are acquired strictly through free news
sources. Whether these data sources differ substantially from paid sources is the subject of future evaluation.
Data Availability
The time interval at which these sites are updated can have critical implications for the efficacy of the public health response. Ideally the source should provide timely reports. In reality, media reporting may be guided by external factors such as a weekly health/science section or unrelated news events that might delay reporting. For example, news volume is strongly affected by day of the week, with high volume on Fridays and low volume on Sundays.
News media reports may also be subject to bias about which diseases are covered. Our evaluation found that the richness of pathogen reporting across news sources was substantial, with 66 unique infectious diseases reported through Google News in the 20-week period (Table 2-1). However, we found that distribution of reports across pathogens (or pathogen evenness) was low, with a substantial skew toward reporting of outbreaks of avian influenza and norovirus. The more skewed distribution in the news sources is expected given the tendency for the media to focus and sustain reporting on stories of public interest. We also
TABLE 2-1 Top Infectious Disease Alerts from the HealthMap System, October 1, 2006–February 16, 2007
|
Disease Reported |
Total Number of Reports |
|
Avian influenza |
661 |
|
E. coli |
492 |
|
Norwalk-like virus |
242 |
|
Salmonellosis |
217 |
|
Influenza |
169 |
|
Dengue fever |
133 |
|
Herpes |
118 |
|
Cholera |
81 |
|
Undiagnosed |
78 |
|
Gastroenteritis |
46 |
|
Pertussis |
52 |
|
Rift Valley fever |
46 |
|
C. difficile |
33 |
|
Staphylococcal disease |
32 |
|
Diarrhea |
29 |
|
Legionellosis |
28 |
|
Tuberculosis |
28 |
|
Malaria |
26 |
|
Chickenpox |
25 |
|
Measles |
25 |
|
SOURCE: http://www.healthmap.org. |
|
found that news outlets often picked up more common seasonal and endemic conditions (e.g., epidemic influenza, dengue, E. coli, Salmonella). This is in contrast to the ProMED system that explicitly avoids reporting on endemic infections—such as tuberculosis and HIV—or vaccine-preventable diseases (Madoff, 2004).
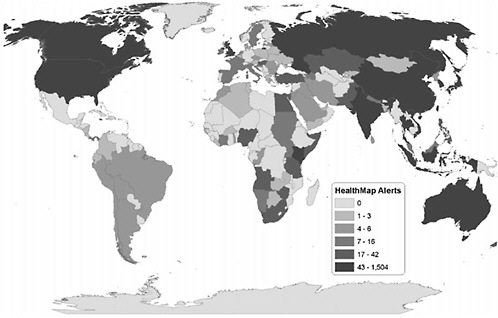
Geographic coverage of data sources also merits quantitative evaluation (Figure 2-5). During the evaluation period, 88 countries had reports of infectious disease outbreaks, with the greatest reporting from the United States (n=1,346), Canada (n=235), and the United Kingdom (n=226). Given that the analysis included only English-language news sources, the skew toward English-speaking countries is not surprising. However, it is also clear there is a bias toward reporting from countries with larger populations (e.g., China), numbers of media outlets, public health resources, and availability of electronic communication infrastructure (approximated by number of Internet hosts).
Future Work in Data Acquisition
Gaps in population and geography covered by news sources need to be understood and adjustments need to be made. For example, important gaps in media reporting exist in tropical areas, which also have the greatest burden of infectious diseases. Monitoring other Internet-based sources such as blogs, discussion sites, and listservs could complement news coverage. In particular, the use of clickstream data and individual search queries is a promising new surveillance source (Eysenbach, 2006). Ultimately, informal news-based sources should be considered as part of a comprehensive multistream surveillance system that provides an integrated view of global health information.
Characterization
Although free and unrestricted websites have large quantities of useful information about infectious diseases, the information is not well organized. News media output usually comes as unstructured free text, making analysis of the geographic and temporal relationships between different reports and data sources difficult. Automated disease and geographic location grouping is usually accomplished through natural language interpretation and automated text mining and parsing. Search criteria can include disease names (scientific and common), symptoms, keywords, and phrases. Once gathered, automated approaches for initial filtering often require human verification.
Classifying Information
Extracting a pathogen name from a free text report presents a number of formidable challenges. In HealthMap, we draw from a continually expanding
dictionary of pathogens (including both human and animal diseases) to organize unstructured and semistructured disease outbreak alert information. Locations are extracted by matching geographic names with a master table of latitude and longitude coordinates of centroids of certain geographic areas, including countries, provinces/states, and cities. In addition to reasonable performance and scalability, a key advantage to this pattern dictionary approach is that it is translated relatively easily to other languages: A different dictionary can be plugged in easily to the existing architecture. A language expert is needed to perform the initial translation, refine the pattern library, help with capitalization and punctuation subtleties, and provide other adaptations, but the basic approach can be re-applied without major changes to the system. Furthermore, the language expert needs to have minimal technical knowledge with respect to natural language syntax or software development to contribute to the library.
Although effective for rapid matching, a number of hurdles need to be overcome. First, in the case where a word may have multiple spellings, for example, diarrhea (common in the United States) and diarrhoea (common in the United Kingdom), we stock the dictionary with multiple patterns for a single pathogen. While look-up time does not increase substantially with the addition of patterns to the dictionary, the disadvantage of the dictionary approach is that it requires a priori knowledge management and allows identification only of locations and diseases already present in the database. Similar challenges exist for identifying the precise geographic location of an outbreak, as geographic names and borders are subject to change. The expansion and editing of the database requires extensive and careful manual data entry. Another limitation of the look-up engine is that it ignores pattern context. A good illustration is the use of “plague” in reports—often news articles use it metaphorically, such as “Problems continue to plague New Orleans cleanup effort.” If the look-up engine matches the word “plague” alone, it will mark this alert as an outbreak of Yersinia pestis in Louisiana. We mitigate this problem by including “to plague,” “a plague,” and other similar strings as null patterns in the library so that the classifier will mask them.
Rating Information
Clearly, the article text contains the best indicators about the locations and diseases of the event in question. However, blindly searching the text, while increasing sensitivity, leads to excessive false positives. To mitigate this problem, we process the input in stages: If the classifier cannot identify location and disease from the initial input provided by the feed, namely the modified headline, it can request more text from the feed. For example, in the case of the Google News aggregator, the system examines the headline, then the description, which usually consists of the first one or two sentences of the article, and finally the publication name. Frequently, a publication in one area refers to events in another area, making the publication name and location an unreliable source for the location
of the alert. However, articles that do not refer to a well-known location, such as “Suburban school closed after flu outbreak,” generally refer to a location near the publication headquarters. By processing the input in stages, we reduce the false positives of the first case while including more of the true positives of the second case.
Future Directions in Data Characterization
Future work must focus on improving natural language processing capability to clearly identify the pathogen, filter nonpertinent reports and duplicates, and enhance the spatial resolution of location. Ideally improvements in how the source information is reported would vastly enhance characterization. For example, structured annotation on the attributes of an outbreak by the article author or source publication would remove the problems inherent with natural language processing. However, given that data standards for news reporting are not likely to be implemented in the short term, advanced text processing methodologies such as fuzzy matching and neural networks could have an important role in enhancing current systems. Furthermore, reliance on an external geocoder with consistently updated databases of geographic locations may be a better solution (Croner, 2003). Although machine learning techniques are undoubtedly important, human analysis still has tremendous value, as exemplified by the Global Public Health Intelligence Network (GPHIN) (Mawudeku and Blench, 2006) and ProMED (Madoff and Woodall, 2005). The success of Wikipedia has shown that leveraging collaborative human networks of trained public health professionals (such as ProMED subscribers and international groups of experts) could be an ideal mechanism for classification, severity assignment, conflict resolution, geocoding, and confirmation of reports on outbreaks of rare or even infectious diseases of unknown identity (Giles, 2005).
Interpretation
While issues of acquisition and characterization have been addressed by many systems, methods for interpreting these data are for the most part underdeveloped. Current systems aggregate Internet-based news resources, but are limited in terms of analytical tools available to the user. Development has been geared toward knowledge management, where news on infectious disease is aggregated and reorganized. Because of the magnitude of information collected, users could, over time, become overwhelmed with an increasing number of false alarms. Thus, there is a need to move from simple knowledge reorganization to an analytic approach for disseminating timely yet specific signals. A number of strategies are available to reduce the false alarm rates in these inherently noisy data sources.
Multistream Interpretation
False alarms often can be reduced by thorough aggregation and cross-validation of reported information on a particular disease outbreak. The motivation for such a meta-alert is based on the idea that multiple sources of information on an incident can provide greater confidence in the validity or reliability of the report than any one source alone. In HealthMap, the severity of a meta-alert is calculated as a composite score based on: (1) the reliability of the data source (e.g., increased weight to WHO reports and less weight to local media reports); and (2) the number of data sources, with increased weight to multiple types of information (e.g., discussion sites and media reports on the same outbreak).
For evaluation of multistream surveillance to be effective, basic characteristics such as sensitivity, specificity, and timeliness of different news source types need to be quantified (Wagner et al., 2001; CDC, 2001; Reis and Mandl, 2003a; Brownstein et al., 2005b; Bloom et al., 2007). In our evaluation, we used officially confirmed outbreaks obtained from WHO Outbreak News, available in the public domain, as a “gold standard” indicator of an infectious disease outbreak (WHO, 2007). We measured key detection characteristics of Google News reports for 12 focused outbreaks over the 20-week period. Mean timeliness for Google News, defined as the time between detection by the surveillance source and report by WHO, was 12 days. However, actual timeliness varied widely from 102 days earlier to 59 days after the WHO report. For example, a diarrheal outbreak in Ethiopia was detected by the media nearly three weeks before the WHO report. In contrast, a plague outbreak in the Democratic Republic of the Congo and a Chikungunya outbreak in India were only reported in the media once the official WHO report was released. Sensitivity, defined as the proportion of WHO alerts detected by news data, was moderate, with 58 percent of the alerts reported in the news. In contrast, we identified 267 unique alerts (country–disease pairs) from Google News, revealing a high volume of reporting. Given that only a subset of outbreaks is posted to WHO Outbreak News, the specificity of news data could not be calculated given the current data sources. Without a better gold standard of validated outbreaks, assessing false positives is difficult.
Statistical Interpretation
The value of news reports can be measured similarly to traditional surveillance data sources used for outbreak detection, where the goal is to distinguish an abnormal pattern from a normal or expected one. Statistical methods for outbreak detection include temporal pattern models such as statistical process control (SPC) (Hutwagner et al., 1997) and autoregressive moving average models (ARIMA) (Reis et al., 2003), spatial models for geographic cluster detection (Kulldorff and Nagarwalla, 1995; Brownstein et al., 2002; Olson et al., 2005), and spatiotemporal patterns for detecting space-time interactions (Kulldorf et
al., 2005). To apply these to monitoring Internet news sources, we must define the baseline patterns and thresholds of reporting for which no action is required. In this case, we expect a baseline level of random noise in news media reports generated by case definitional issues, surveillance bias, and overreporting. The generation of statistical signals therefore can be based on a set threshold of report volume defined by modeling this baseline. Models can include factors such as cyclical patterns (day of week, month, seasonal effects) and autocorrelation (Reis and Mandl, 2003b; Brownstein et al., 2005a), as well as geographic and temporal biases of news reporting. Thresholds would be set by evaluating the tradeoff between signal quality and timeliness.
Future Directions in Data Interpretation
Future work in modeling and data integration should also be directed at improving risk assessment. For example, signals from unstructured online information sources can be integrated with other health indicator data to provide a broader context for the alert. Pertinent datasets include mortality and morbidity estimates, population density and mobility, and pathogen seasonality and transmissibility (Wilson, 1995; Altizer et al., 2006; Dowell and Ho, 2004; Grassly and Fraser, 2006; Fraser et al., 2004). With the increasing importance of vectorborne and zoonotic diseases (Gratz, 1999; Dobson and Foufopoulos, 2001; Brownstein et al., 2004), consideration also should be given to inclusion of ecological data such as distribution of arthropod vectors and animal host reservoirs, as well as environmental predictors including climate and vegetation (Brownstein et al., 2003; Colwell et al., 1998; Kitron, 1998). Combining these informal sources with clinical and laboratory surveillance data should also be an important next step. Such integration could yield a relevancy score for the report, define populations at risk, and predict disease spread.
Dissemination
An important final consideration is how information from Internet-based systems should be disseminated. Clearly a critical audience is public health officials interested in real-time updates of infectious disease status in their geographic region. However, whether these systems should be freely available and open to the public is an area of active debate. Travelers, for example, may have a keen interest in up-to-the-minute knowledge about infectious disease activity at their destination. For the general population, obtaining integrated real-time coverage of a disease emergency is particularly challenging given disparate news and alert sources. An unrestricted sentinel system dedicated to the aggregation and geographic display of current outbreaks could fill this information gap.
On the other hand, unrestricted access to this information could have severe
economic impacts on the countries affected by the disease alerts. The risk is especially elevated with fully automated systems that may not have the benefit of informed human judgment. However, even with careful controls, any system can generate spurious alerts. User restrictions may be the only way to guard against unwarranted damage (Cash and Narasimhan, 2000). Furthermore, an open access model might not be economically sustainable. GPHIN depends on subscription fees to make any necessary improvements to the system. In contrast, HealthMap is based on freely available data and is open to the public. However, a tiered approach such as the one used by MedISys (Medical Intelligence System) where general information is provided to the public based on free resources and more detailed information pertinent to public health officials (including geographic detail) is provided by paid subscription may represent a reasonable compromise. Information access is a key consideration for the future development of these systems.
Another critical question is who ultimately should oversee these systems and manage the information collected, especially given that issues of trust and reliability are paramount. At the moment, systems are being developed by international organizations, governments, and academic institutions. The current linkage between surveillance by GPHIN and public health response by WHO’s GOARN presents a very appealing approach. An initiative at Google.org, called the International System for Total Early Disease Detection (INSTEDD) project, aims to develop a system that increases the number of languages and data sources available through GPHIN (Delamothe, 2006). INSTEDD could become a transparent and publicly available resource independent of any government agency. How such a system would be linked with existing public health infrastructure should be an area of active consideration.
Conclusions
The growing use of informal electronic information sources highlights an important paradigm shift in disease surveillance, expanding beyond traditional public health systems. Although Internet-based informal sources on outbreaks are becoming a critical tool for global infectious disease surveillance, important challenges still need to be addressed. In particular, an unavoidable pitfall of a system-of-systems approach is that it is inherently subject to the limitations of the primary data collected by the individual component systems. Our preliminary evidence-based evaluation of HealthMap suggests that aggregation of multiple sources may counter this limitation by increasing sensitivity and timeliness while reducing false alarms, in that assessments are not based on any single news outlet alone. Because many of the places with the least technological adoption also carry the greatest infectious disease burden, future system development should also specifically address the digital divide to achieve more uniform and comprehensive global coverage. A robust research agenda, continued organic evolution of existing and new technologies, and scrutiny through a rigorous evaluation framework
will help ensure that the global public health enterprise maximally leverages new electronic sources for surveillance, communication, decision making, and intervention.
Acknowledgments
This work was supported by R21LM009263-01 and R01LM007970-01 from the National Library of Medicine, the National Institutes of Health, and grant number 200510MFE-154492 from the Canadian Institutes of Health Research.
USING CELL PHONE TECHNOLOGY FOR INFECTIOUS DISEASE SURVEILLANCE IN LOW-RESOURCE ENVIRONMENTS: A CASE STUDY FROM PERU
Pamela R. Johnson, Ph.D.15
Voxiva
David L. Blazes, M.D., M.P.H.16
Naval Medical Research Center Detachment
Summary
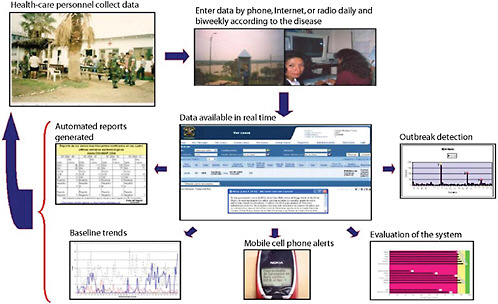
Basic routine health surveillance has been largely unachievable in most of the developing world because of inadequate funding and public health training, especially in resource-limited parts of Africa, Asia, and Latin America. Considering that many emerging diseases with pandemic potential first occur in the developing setting (e.g., severe acute respiratory syndrome, or SARS and H5N1 avian influenza), enhanced surveillance systems in these countries must become high priorities for safeguarding global public health. This presentation reports on an innovative model using a cell phone- and Internet-based reporting system that has been developed and tested to extend disease surveillance by the Peruvian Navy along the coast and remote rivers of Peru. Alerta DISAMAR—the name given to the system refers to the Health Department of the Peruvian navy—has been fully operational in Peru since 2003. More than 600 individuals have been trained and have used an Internet- and cell phone-based electronic system developed by Voxiva to report routinely from 42 land-based sites and 19 ships. More than 80,000 cases and 31 outbreaks have been reported. Alerta DISAMAR represents a sustained, large-scale effort that leverages cell phones and related tech-
nological innovations to strengthen disease surveillance. Because mobile phone networks are expanding so rapidly across the developing world, this model has important lessons for strengthening disease surveillance in other low-resource environments.
Background
The low-resource environments of most developing countries pose a particular challenge to global efforts to establish effective infectious disease surveillance and detection. There is little question that these countries are important to global surveillance. “Out of every 100 persons added to the population in the coming decade,” Zlotnik wrote, “97 will live in developing countries” (Zlotnik, 2005). Figure 2-6 shows countries sized in proportion to their populations. It shows how significant the share of the world’s population is in China, Brazil, Nigeria, sub-Saharan Africa, and other countries in the developing world.
Infectious disease is disproportionately represented in these countries. Many new and emerging infectious diseases—including SARS, H5N1, HIV/AIDS—trace their origins to these often densely populated environments. However, despite their demographic and epidemiologic significance, most developing countries, where microbial threats to global health are most likely to emerge, also possess the weakest surveillance systems. The Government Accountability Office (GAO), in a global review of surveillance systems, noted that:
Developing country systems are a weak link in the global surveillance framework. Surveillance systems in industrialized and developing countries suffer from a number of common constraints, including a lack of human and material resources, weak infrastructure, poor coordination, and uncertain linkages between surveillance and response. However, these constraints are more pronounced in developing countries, which bear the greatest burden of disease and are where new pathogens are more likely to emerge, old ones to reemerge, and drug-resistant strains to propagate. Weaknesses in these countries thus substantially impair global capacity to understand, detect, and respond to infectious disease threats (GAO, 2001).
Over the past decade, a number of important efforts have been made to use information technology to strengthen surveillance systems. Most have taken place in developed countries where computers and Internet connectivity are readily available. In addition, e-mail and the Internet have had a major impact in facilitating the growth of global networks such as the World Health Organization’s (WHO’s) Global Outbreak Alert and Response Network (GOARN), ProMED-mail, and a number of global disease-specific surveillance networks (Heymann and Rodier, 2004).
Despite the evident importance of information technology (IT) in these
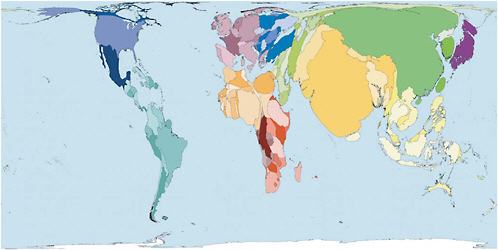
FIGURE 2-6 Distribution of the earth’s population.
SOURCE: Worldmapper (2007). See http://www.worldmapper.org. Reprinted with permission. © Copyright 2006 SASI Group (University of Sheffield) and Mark Newman (University of Michigan).
examples, IT has yet to realize its full potential for strengthening the surveillance systems of countries in the developing world. Even where computers exist, inadequate power and lack of Internet coverage outside major towns and cities has meant that in most developing countries routine surveillance systems are still largely paper based, and transmission is slow. Health officials in capital cities and towns may be easily linked to global networks and colleagues in other major cities, but communicate less easily with health facility staff in their own countries.
Until recently, there was no practical, measurable way to bridge the digital divide between the cities and the poor and rural areas in these countries, where most of the population lives. Nonetheless, a major recent review of disease priorities led by The World Bank holds out a vision of the future of disease surveillance:
Public health agencies, ministries of finance, and international donors and organizations need to transform surveillance from dusty archives of laboriously collected after-the-fact statistics to meaningful measures that provide accountability for local health status or that deliver real-time early warnings for devastating outbreaks…. Information technology and informatics can help in attaining this vision … technology can facilitate the collection, analysis, and use of surveillance data, if data standards are developed and compatible systems are established. … technology such as cell phone-based systems could accelerate collection of key data (for example, occurrence of a viral hemorrhagic fever outbreak) (Nsubuga et al., 2006).
The explosive growth of mobile telephone networks in the developing world is already a dramatic success story. According to the International Telecommunications Union (ITU), Internet connectivity is growing at a good pace, but the reality is that cell coverage is growing much faster. In the developing world there are 1.2 billion phones and a million new mobile phone subscribers every day. Approximately 80 percent of people who live today are within reach of a mobile phone signal, according to the ITU. Half of all households will have phone access in the next decade, and 90 percent of the world will be covered by 2010. In Latin America, there are more than 270 million mobile phone users today. The industry estimates that within the next 3 years, 75 percent of people in Latin America will own a cell phone. Africa shows the most dramatic growth. Over the past 5 years, growth has averaged nearly 60 percent a year, with nearly 76 million subscribers at the end of 2004.
Voxiva was founded in 2001 to find practical ways of using information technology for health and development in low-resource environments. We reviewed a variety of pilot projects that were seeking to extend the benefits of the Internet using a variety of individual devices, including personal digital assistants (PDAs) and satellite dishes. However, we did not find any practical, measurable, and sustainable strategy to support data collection and communication with points
of service. It was and in many countries remains a paper-based world, with few benefits of IT reaching large scale.
Voxiva’s founders recognized the potential of the growth of cellular networks to develop a measurable, sustainable approach that could support public health and development. Working with a variety of public health agencies, including health officials in the U.S. and Peruvian navies, Voxiva developed innovative software that allows health professionals to enter and access data using the Internet, a cell phone, or other devices. The software makes it possible to write a survey and to make that survey available in multiple formats so users can use the tools they have and the most convenient, cost-effective means to respond.
Figure 2-7 illustrates the approach that Voxiva has taken to optimize the use of the existing infrastructure of personal computers (PCs), Internet, cell phones, fixed phones, pay phones, PDAs, smart phones, and paper to create networked data collection and communications surveillance systems in low-resource environments.
Over the past six years, we have learned a number of lessons for building IT systems for use in low-resource environments, leveraging cell phones and other forms of information technology.
Building on Available Infrastructure
The global mobile phone network is increasingly the most important globally deployed communications infrastructure that covers the developing world. As fast growing and highly successful local businesses, telecommunications companies provide global infrastructure for data transmission and communication that is inherently sustainable. As a result, the health sector no longer needs to build and maintain its own infrastructure to transmit data and support communication with its network of health facilities.
Hardware is also increasingly available. Health ministries, states or provinces, and increasingly districts have computers and at least intermittent Internet access. In addition, the dramatic expansion of mobile phone usage has put a simple “terminal” within the reach of many if not most health workers—one that can be used to enter data, respond to surveys sent by text messages, and send and receive alerts. Providing a toll-free number that people can call with their own mobile phones is a quicker and cheaper approach to expanding a surveillance network than buying, equipping, maintaining computers, and paying for power and Internet access for all health facilities.
By building on this global telecommunications infrastructure and available hardware, it is possible to rapidly create large-scale integrated networks that can do basic reporting without large-scale new investments in acquiring, installing, and maintaining technology. Over time, the infrastructure will grow—but most countries already have enough hardware to begin.
As one example, Voxiva was able to support the national scale-up of
Rwanda’s national HIV/AIDS program with a monitoring system that allows health facilities to report program indicators and register new patients. Without any new investment in hardware, Rwanda was able to rapidly increase its electronic reporting system from 12 facilities to 146 sites. Ninety percent of the sites report program indicators and register patients using the phone. They use their own cell phones, call a toll-free number—the first in Rwanda—log in using their password, and enter basic data digitally using the keypad. Over time, more and more sites will get computers and Internet connectivity, but the program was able to reach national coverage rapidly without waiting.
Multiple, Redundant Technologies
We have found that no single solution or hardware works in all environments, for a variety of reasons. Power is a major constraint, especially in the most remote environments and poorest countries. The cost of connectivity remains high in many countries. Maintenance and support for computer hardware and software are costly and in short supply. Actual needs for bandwidth and analytic power vary depending on how users are using the data collected. For many needs, paper records will be the base system. Having the choice of different electronic devices means that one can optimize the use of available hardware and allows health professionals to enter and access data using the access means they have. This also creates redundancy: If the Internet is not available, it is still possible to do basic reporting via the phone.
Electronic Data Capture at the Source
Paper systems rely on copying information at the source, relaying it, then entering and aggregating information at higher levels; this system of data transmission is error prone and makes it difficult to trace data and cases back to their source. If data can be entered and confirmed at the source, this in itself can enhance quality. If data can be transmitted to a central database, reviewed and approved at higher levels (e.g., at the district level), and also be made rapidly available to others who need it, the burden and risks associated with entering data multiple times are reduced.
Rapid Transmission of Data
Most surveillance systems rely on physical transmission of data via mail or “sneaker net” with people bringing the forms to a monthly meeting or on a periodic visit. More urgent information is transmitted via individual phone or radio calls and recorded centrally. Electronic transmission data can cut the time and cost required for transmission dramatically. Although this is important for routine surveillance, it is even more vital for an event, such as a serious outbreak
or pandemic, when the situation on the ground could change rapidly. Electronic submission of data in near real-time information could help decision makers make much better decisions about allocation of resources in a situation of rapid change.
Shared Database, Role-Based Access
If data are collected at the source and transmitted to a core database, organizations can make that data available to authorized users according to the specific roles they play. For example, the same set of data could be viewed in a variety of ways:
-
Health staff can get confirmation that their report has been received, notification of cases of interest from neighboring locations, and results of a case investigation;
-
A district health official could see reports from health clinics and posts immediately—then edit or approve them;
-
Senior health officials could review aggregate data;
-
Surveillance officers could get lists of nonreporting sites; and
-
Outbreak investigation teams or vaccination teams could get short message service (SMS) or e-mail alerts about suspected cases of measles or potential outbreaks.
Data, collected once and transmitted to a central database, can be used many times. Furthermore, data can be presented in standard templates, basic reports, and maps or exported for additional analysis.
Communication Plus Feedback
An extended electronic network can also facilitate communication and feedback that is vital to the supervision and motivation of a distributed network. Automated messages can be sent by multiple means—e-mails, text messages, voice mails, alerts—and accessed through the available technology.
Build Human Capacity
Information technology alone is no silver bullet. Building a surveillance system in any environment requires an investment in the training staff at all levels. Well designed information systems can help, with tools such as validation rules, reminders, and online access to guidelines and training materials. Such tools could be even more helpful in situations where health staff are not well trained. However, the best designed system cannot detect disease or respond to
an outbreak without the right processes and people; at its core, a surveillance system will always be about the people who use it. The role of technology should be to empower networks of clinicians, nurses, and other health workers to fully participate in this important enterprise of global disease surveillance.
The same technological approach leveraging cell phone technology is being used for a variety of purposes. It has been used for syndromic surveillance in schools in San Diego and Washington, DC. It has been tested in the Canete Valley in Peru and in Baghdad and Basra, Iraq, and is being developed with the Ministry of Agriculture in Indonesia to create a system to strengthen animal surveillance. In Latin America, it is being used to monitor national HIV/AIDS programs, track adverse events, and support public safety (Olmsted et al., 2005; Curioso et al., 2005).
Alerta DISAMAR: A Case Study in Infectious Disease Surveillance
Background
In fall 2001, at a time when other events in the world raised awareness of microbial and other threats, there was an outbreak of P. falciparum malaria at a remote naval base in the Amazon jungle basin of Peru that led to several deaths. As in some other countries, the existing surveillance system was underdeveloped and primarily paper based; it was not unusual for reports to take a month to get to authorities in Lima.
Primed by these events, the Peruvian navy, the U.S. Naval Medical Research Center Detachment in Lima, and the Peru-based office of Voxiva developed a joint project. In January 2003, they initiated Alerta DISAMAR, a novel electronic disease surveillance system. This experience in implementation has been exciting to observe from the ground up, and this case study describes some of the lessons learned during deployment of this surveillance system in Peru.
Pandemics by definition involve the global dissemination of disease. Military populations historically have been involved in the dissemination of a number of infectious diseases, including their well-documented role in the spread of H1N1 influenza in 1918–1919 (Oxford et al., 2005). There are also examples from antiquity of troop movement roles in the spread of smallpox, cholera, measles, syphilis, and plague (McNiel, 1977). More recently, high rates of HIV infection have been seen in sub-Saharan African militaries, and this population certainly contributes to ongoing transmission (Whiteside and Winsbury, 1996). Despite the known risks of disease transmission among highly mobile armed forces, these groups remain among the most poorly tracked populations in many developing nations. The combination of undersurveilled military populations and dangerous transmissible diseases seems the perfect recipe for a pandemic.
Military personnel are a very good population to place under surveillance for
emerging infectious diseases (Chrétien et al., 2007). They serve as ideal sentinel populations due to their expeditionary mission, their frequent travel to remote locations, and their interactions with local populations.
A number of challenges exist in implementing an electronic disease surveillance system, some cosmopolitan and some unique to resource-limited settings. The dubious observer may even ask if conducting surveillance in a resource-limited setting is even feasible when the potential pitfalls and challenges often seem insurmountable. The first challenge is to create a system that is complementary and not duplicative of existing surveillance systems. Creation of parallel surveillance systems can paradoxically lead to the failure of both the established and the new systems because limited funds and effort can be diluted between the two systems with neither functioning effectively (Nsubuga et al., 2006). Second, one must convince the stakeholders in the population under surveillance that their efforts are useful and their valuable time is not wasted. To accomplish this, one must make sure that meaningful data are returned to the end users of the system, and in a timely fashion so that consequential action may be taken to limit the effect of a disease or condition. An example might be the timely detection of an influenza outbreak in a closed facility where case isolation, enforcement of hand/cough hygiene, and antiviral chemoprophylaxis may limit the spread of this contagious illness.
Additionally, a disease surveillance system in the developing setting must be cost-effective, with few recurring expenses in order to assure sustainability. Many countries in the developing world have limited funds to spend on public health, and these are often exhausted in responding to crises rather than invested in preventive strategies. Finally, a disease surveillance system in the developing setting must be able to function in remote locations and austere conditions.
With regard to the situation in the Peruvian navy, we focused our attention on a population that was not under surveillance, and applied novel technology in the attempt to create a model that could be disseminated to other resource-limited settings worldwide. Furthermore, in establishing this surveillance system, we have attempted to change the culture within this population to approach disease characterization and transmission from a broader perspective, that of epidemiology and public health.
The System
The Alerta electronic disease surveillance system uses a countrywide network of health-care facilities that encompasses more than 95 percent of the population of the Peruvian navy and its civilian dependents in most regions of Peru (over 120,000 people). Some of these sites are tertiary care facilities in the capitol city of Lima, but the majority are smaller, more remote, and less capable clinics that exist throughout the country. Figure 2-8 shows a typical clinic site in one of the austere areas where this system functions.

FIGURE 2-8 Health-care personnel collect data.
SOURCE: Jose Quispe, Peruvian Navy. Reprinted with permission from NMRCD.
The network consists of 43 fixed sites throughout Peru and 19 ships both on the coast and on rivers in the jungle (Figure 2-9). The sites use a diverse range of reporting techniques, with 12 sites routinely reporting by radio relay, 27 by telephone, 15 by Internet, and 8 by telephone or Internet.
Data flow from individual clinics to the central operations hub in Lima. The actual data flow from the field is described in Figure 2-10. Data are collected on standard clinical forms during patient encounters, then entered into the system by nurses or physicians via cell phones, toll-free public telephones, or by Internet if accessible. Several extremely remote sites are beyond the cellular footprint, and in these, the Peruvian navy personnel use a radio phone to relay data to the next nearest site that has either a cell phone or access to public phones or the Internet. Occasionally, satellite phones are used if they are available to enter data through the toll-free telephone system.
The data collected through this system are often the only systematic representation of the epidemiology of diseases within the Peruvian navy. This has allowed baseline levels of disease to be set, and allocation of scarce resources to be assigned based on rational data rather than assumptions. The system collects rates of the 29 reportable illnesses for the Peruvian Ministry of Health (MoH) as well as some militarily relevant cases such as training-related injuries. The data generated by this system are invaluable to both the Peruvian navy and the MoH
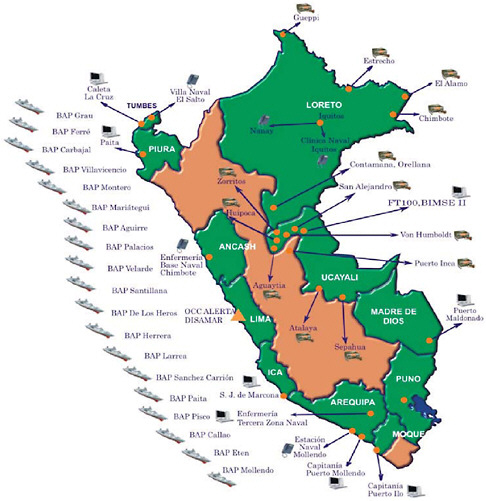
FIGURE 2-9 Alerta network.
SOURCE: Dr. Carmen Mundaca, Naval Medical Research Center Detachment (NMRCD). Reprinted with permission from NMRCD.
because they report disease rates from areas of the country where the MoH does not have a significant presence.
The data are captured and displayed in real time on a web-based platform. Several automated outputs are generated so that feedback is given almost immediately to the stakeholders in this process, either by electronic mail or short message service (SMS) messaging to cellular phones. Features include automated outbreak detection via algorithms, graphical representation to assist clinicians, and baseline trends.
Discussion
A complete evaluation of the ALERTA surveillance system was performed for the period from January 2003 to November 2006, and is the subject of a separate publication (Mundaca et al., 2005). This evaluation focused on three phases: implementation (first year), consolidation (second year), and expansion (third and fourth years). The methods for this evaluation are based on the Centers for Disease Control and Prevention’s (CDC’s) Updated Guidelines for evaluating public health surveillance systems (CDC, 2001). The tasks involved in evaluating this system are adapted from the steps in program evaluation in the Framework for Program Evaluation in Public Health (CDC, 1999), as well as from the elements in the original guidelines for evaluating surveillance systems (CDC, 1988). This assessment was based on information from several data sources, including the main database generated by the system platform, quarterly morbidity reports from the Peruvian navy, outbreak reports, information from Voxiva personnel, focus groups, training evaluations, and surveys applied to stakeholders. Highlights of this evaluation are included below, and include usefulness, sustainability, stability, and flexibility.
The Alerta system has been invaluable to the Peruvian navy. Since its implementation through November 2006, 80,747 events have been reported, including 3,789 in 2003; 9,454 in 2004; 25,246 in 2005; and 42,258 through November 2006. The Peruvian navy has embraced Alerta DISAMAR and the culture of epidemiology surrounding it. As one example, the Peruvian military leadership asked all the services for the number of cases of dengue fever in the past year. The navy was the only group that could provide a number and distribution within the week. They searched Alerta DISAMAR’s database and were able to provide the information rapidly. Since this incident, the other branches of the Peruvian military have decided to implement Alerta. Reports such as these have allowed baseline levels of disease to be determined, and for the first time have identified outbreaks of disease in a timely fashion so that diagnoses can be made and interventions enacted.
One of the most important questions to ask in evaluating a system is whether that system is doing what it was intended to do. Over the past four years, we have detected more than 31 outbreaks, including diarrhea, dengue, influenza, and tuberculosis. The outbreak of diarrhea depicted below is an example of an outbreak reported using the system (Figure 2-11), and there have been several outbreaks of acute respiratory infections that have initiated outbreak responses at recruit training camps. One of these identified outbreaks (mumps) led the Ministry of Health (MoH) to conduct active community surveillance that uncovered an ongoing outbreak in the civilian population that mirrored that found in the Peruvian navy.
Timely detection of outbreaks of disease allow accurate laboratory diagnoses to be made, and with a firm diagnosis, a viable response can be fashioned
that hopefully will attenuate the outbreak. The Naval Medical Research Center Detachment (NMRCD) has been able to marry Alerta to molecular and microbiologic diagnostics in a number of these outbreaks, including the use of rapid antigen detection testing for influenza.
In addition to outbreak response, we have also provided training in basic epidemiology and more advanced field epidemiology. We have trained more than 600 public health personnel in the Peruvian navy in basic epidemiology and the use of this electronic disease surveillance system. Throughout South America, we have also trained more than 1,300 epidemiologists in the basics of outbreak detection and management. The objectives for these courses and the entire curriculae in Spanish and English are available at no cost on the Web (Lescano et al., 2007).
The following attributes of the Alerta system were included in the evaluation process:
-
Simplicity: Description of the data flow; estimated time for the reporter to collect information and analyze the data; staff training requirements; and time spent on the maintenance of the electronic platform.
-
Flexibility: Number of reporting sites added per year; cost and time required to add new sites; ability to add new diseases to the reporting template.
-
Data quality: Reporting rate (percentage of sites that report per total number of sites); percentage of complete reports; error rate (number of errors/ number of reports); error rate per site (number of errors/total number of sites per week).
-
Acceptability: Personnel surveys after training courses; number of personnel who report per site; mean time after training to achieve a timely report.
-
Representativeness: Coverage (percentage of Naval population covered by the system); characteristics of the population.
-
Timeliness: Percentage of sites that report on time and percentage of outbreaks detected on time; average of days to report.
-
Stability: Number of system failures; percentage of time that the system is fully operational; actions involved with repairs in the system.
-
Sustainability: Joint responsibilities; relationship with the Peruvian navy; incentives; costs assumed by each part; problems and requirements to sustain the system.
Overall, the Alerta electronic disease surveillance system has been embraced by the Peruvian navy and has transformed public health preparation and response in this population. Both the Peruvian navy and the NMRCD laboratory have contributed personnel, resources, and significant time to ensure optimal performance. The implementation of this system has not been without pitfalls, and many challenges persist. However, the significant progress illustrates how horizontal partnerships and small projects can generate measurable improvements in epidemiologic capability.
This quote from a U.S. navy physician sums up the experience in Peru:
The introduction of Alerta has led to early outbreak identification/response, timely case management, and increased review of clinical procedures within reporting units… [It is a] working model for similar larger scale international programs Alerta is a simple, near real-time disease surveillance model for countries in all stages of communications technology development (Lescano et al., 2003).
Clearly, to respond to and control a potential pandemic, all regions of the world need fully functional public health systems. These systems require careful networking of many components, including reliable disease surveillance, accurate local diagnostics, rapid medical response capability, and fluid cooperation and communication among local and international partners.
Some components of successful public health strategies are present in the U.S. Department of Defense Global Emerging Infections Surveillance and Response System (DoD-GEIS). This system is a decade-old DoD program initiated in response to President Clinton’s directive in 1996 that mandated the development of a global system to track, control, and respond to potential pandemic infections. It generated, among other things, the electronic disease surveillance system described above (White House, 1996). GEIS serves as just one component of a growing network of public health assets that are increasingly being used to control infectious diseases with pandemic potential, complementing many global public health community efforts (Chrétien et al., 2006).
The Alerta model implemented in Peru has a number of dimensions that have contributed to its success:
-
Committed leadership in all parties;
-
A regulatory regime that specified reporting requirements;
-
A practical use of information technology that maximized the use of available telecommunications and computing infrastructure;
-
Real-time data collection from points of service and automated reports and notification;
-
Live database for continuous analysis and investigation;
-
Links to laboratory and investigation capacity;
-
Training and support of a distributed network of clinicians and other health workers; and
-
Mobile technology accessible to virtually everybody in Peru—if not individually then through a Navy command with cell phones and Internet access.
The approach that was developed and tested in Peru is now being expanded with support from the U.S. DoD Southern Command to five neighboring countries: Bolivia, Colombia, Ecuador, Paraguay, and Uruguay. In addition, Voxiva is part of a public–private partnership with the GSM Association, the largest asso-
ciation of mobile phone operators (more than 650 mobile phone operators and 2 billion subscribers worldwide) to extend the benefits of this network in service to public health. Hopefully this can provide not only a model of working in the field, but also a model of cooperation between public and private entities.
Acknowledgments
The results reported derive from the common vision and hard work of a number of individuals in the Peruvian and U.S. navies and Voxiva as well as the support of the DoD-GEIS program. Key individuals include Drs. Carmen Mundaca, Roger Araujo, Tanis Batsel, Rafael Elgegren, Ernesto Gozzer, Patrick Kelley, Andres Lescano, Monica Negrete, and Mario Ortiz.
REFERENCES
Altizer, S., A. Dobson, P. Hosseini, P. Hudson, M. Pascual, and P. Rohani. 2006. Seasonality and the dynamics of infectious diseases. Ecology Letters 9(4):467-484.
Baker, M. G., and D. P. Fidler. 2006. Global public health surveillance under new international health regulations. Emerging Infectious Diseases 12(7):1058-1065, http://www.cdc.gov/ncidod/EID/vol12no07/pdfs/05-1497.pdf (accessed May 15, 2007).
Bloom, R. M., D. L. Buckeridge, and K. E. Cheng. 2007. Finding leading indicators for disease outbreaks: Filtering, cross-correlation, and caveats. Journal of the American Medical Informatics Association 14(1):76-85.
Brownstein, J. S. 2006. HealthMap: Global Disease Alert Mapping. Presentation at the Institute of Medicine Forum on Microbial Threats, Washington, DC, December 12-13.
Brownstein, J. S., H. Rosen, D. Purdy, J. R. Miller, M. Merlino, F. Mostashari, and D. Fish. 2002. Spatial analysis of West Nile virus: Rapid risk assessment of an introduced vector-borne zoonosis. Vector Borne Zoonotic Diseases 2(3):157-164.
Brownstein, J. S., T. R. Holford, and D. Fish. 2003. A climate-based model predicts the spatial distribution of the Lyme disease vector Ixodes scapularis in the United States. Environmental Health Perspectives 111(9):1152-1157.
Brownstein, J. S., T. R. Holford, and D. Fish. 2004. Enhancing national West Nile virus surveillance. Emerging Infectious Diseases 10(6):1129-1133.
Brownstein, J. S., K. P. Kleinman, and K. D. Mandl. 2005a. Identifying pediatric age groups for influenza vaccination using a real-time regional surveillance system. American Journal of Epidemiology 162(7):686-693.
Brownstein, J. S., K. L. Olson, K. P. Kleinman, and K. D. Mandl. 2005b. Effect of site of care and age on timeliness and accuracy of syndromic surveillance data [Abstract]. Morbidity and Mortality Weekly Report 54(Suppl):184.
Buehler, J. W., R. S. Hopkins, J. M. Overhage, D. M. Sosin, and V. Tong. 2004. Framework for evaluating public health surveillance systems for early detection of outbreaks: Recommendations from the CDC Working Group. Morbidity and Mortality Weekly Report 53(RR-5):1-11.
Butler, D. 2006. Disease surveillance needs a revolution. Nature 440(7080):6-7.
Captain, S. 2006. Get your daily plague forecast. Wired News, http://www.wired.com/science/discoveries/news/2006/10/71961/ (accessed April 23, 2007).
Cash, R. A., and V. Narasimhan. 2000. Impediments to global surveillance of infectious diseases: Consequences of open reporting in a global economy. Bulletin of the World Health Organization 78(11):1358-1367.
CDC (Centers for Disease Control and Prevention). 1988. Guidelines for evaluating surveillance systems. Morbidity and Mortality Weekly Report 37(S-5):1-18.
CDC. 1999. Framework for program evaluation in public health. Morbidity and Mortality Weekly Report 48(RR-11):1-40, http://www.cdc.gov/mmwr/preview/mmwrhtml/rr4811a1.htm (accessed May 14, 2007).
CDC. 2000. Biological and chemical terrorism: Strategic plan for preparedness and response. Recommendations of the CDC Strategic Planning Workgroup. Morbidity and Mortality Weekly Report 49(RR-4):1-26.
CDC. 2001. Updated guidelines for evaluating public health surveillance systems: Recommendations from the guidelines working group. Morbidity and Mortality Weekly Report 50(RR13):1-35, http://www.cdc.gov/mmwr/preview/mmwrhtml/rr5013a1.htm (accessed May 14, 2007).
Chrétien, J. P., J. C. Gaydos, J. L. Malone, and D. L. Blazes. 2006. Global network could avert pandemics. Nature 440(7080):25-26.
Chrétien, J. P., D. L. Blazes, R. L. Coldren, M. D. Lewis, C. C. Mundaca, J. Green, S. Montano, G. J. Martin, and J. Gaydos. 2007. The importance of developing country militaries in global infectious disease surveillance. Bulletin of the World Health Organization 85(3):174-180.
Colwell, R. R., P. R. Epstein, D. Gubler, N. Maynard, A. J. McMichael, J. A. Patz, and R. Shope. 1998. Climate change and human health. Science 279(5353):968-969, http://www.sciencemag.org/cgi/content/full/279/5353/963g/ (accessed April 23, 2007).
Cowen, P., T. Garland, M. E. Hugh-Jones, A. Shimshony, S. Handysides, D. Kaye, L. C. Madoff, M. P. Pollack, and J. Woodall. 2006. Evaluation of ProMED-mail as an electronic early warning system for emerging animal diseases: 1996 to 2004. Journal of the American Veterinary Medical Association 229(7):1090-1099.
Croner, C. M. 2003. Public health, GIS, and the Internet. Annual Review of Public Health 24:57-82.
Curioso, W. H., B. T. Karras, P. E. Campos, C. B. King, K. Holmes, and A. M. Kimball. 2005. Design and implementation of cell-PREVEN: A real-time surveillance system for adverse events using cell phones in Peru. American Medical Informatics Association Annual Proceedings. Pp. 176-180.
Delamothe, T. 2006. TED 2006: The future we will create. British Medical Journal 332(7540), http://www.bmj.com/cgi/content/full/332/7540/DC1?ehom/ (accessed April 23, 2007).
Dobson, A., and J. Foufopoulos. 2001. Emerging infectious pathogens of wildlife. Philosophical Transactions of the Royal Society of London Series B Biological Sciences 356(1411):1001-1012.
Dowell, S. F., and M. S. Ho. 2004. Seasonality of infectious diseases and severe acute respiratory syndrome—what we don’t know can hurt us. Lancet Infectious Diseases 4(11):704-708.
Eysenbach, G. 2006. Infodemiology: Tracking flu-related searches on the web for syndromic surveillance. Proceedings of the American Medical Informatics Association Annual Symposium. Pp. 244-248.
Formenty, P., C. Roth, F. Gonzalez-Martin, T. Grein, M. Ryan, P. Drury, M. K. Kindhauser, and G. Rodier. 2006. [Emergent pathogens, international surveillance and international health regulations (2005)]. Médicine et Maladies Infectieuses 36(1):9-15.
Fraser, C., S. Riley, R. M. Anderson, and N. M. Ferguson. 2004. Factors that make an infectious disease outbreak controllable. Proceedings of the National Academy of Sciences 101(16):6146-6151.
GAO (General Accounting Office). 2001 (August). Global health: Challenges in improving infectious disease surveillance systems. GAO-01-722. P. 16, http://www.gao.gov/new.items/d01722.pdf (accessed May 14, 2007).
Giles, J. 2005. Internet encyclopaedias go head to head. Nature 438(7070):900-901.
Grassly, N. C., and C. Fraser. 2006. Seasonal infectious disease epidemiology. Proceedings Biological Sciences 273(1600):2541-2550.
Gratz, N. G. 1999. Emerging and resurging vector-borne diseases. Annual Review of Entomology 44:51-75.
Grein, T. W., K. B. Kamara, G. Rodier, A. J. Plant, P. Bovier, M. J. Ryan, T. Ohyama, D. L. Heymann. 2000. Rumors of disease in the global village: Outbreak verification. Emerging Infectious Diseases 6(2):97-102.
Hardiman, M. 2003. The revised International Health Regulations: A framework for global health security. International Journal of Antimicrobial Agents 21(2):207-211.
Henderson, D. A. 1993. Surveillance systems and intergovernmental cooperation. In Emerging Viruses, edited by S. S. Morse. New York: Oxford University Press. Pp. 283-289.
Heymann, D. L., and G. R. Rodier. 1998. Global surveillance of communicable diseases. Emerging Infectious Diseases 4(3):362-365.
Heymann, D. L., and G. R. Rodier. 2001. Hot spots in a wired world: WHO surveillance of emerging and re-emerging infectious diseases. Lancet Infectious Diseases 1(5):345-353.
Heymann, D. L., and G. Rodier. 2004. Global surveillance, national surveillance, and SARS. Emerging Infectious Diseases 10(2):173-175.
Holden, C. 2006. Netwatch: Diseases on the move. Science 314(5804):1363d.
Hutwagner, L. C., E. K. Maloney, N. H. Bean, L. Slutsker, and S. M. Martin. 1997. Using laboratory-based surveillance data for prevention: An algorithm for detecting Salmonella outbreaks. Emerging Infectious Diseases 3(3):395-400.
IOM (Institute of Medicine). 1992. Emerging infections: Microbial threats to health in the United States. Washington, DC: National Academy Press.
IOM. 2003. Microbial threats to health: Emergence, detection and response. Washington, DC: The National Academies Press.
IOM. 2004. Learning from SARS: Preparing for the next disease outbreak. Washington, DC: The National Academies Press. Pp. 4-6.
Jebara, K. B. 2004. Surveillance, detection and response: Managing emerging diseases at national and international levels. Revue Scientifique et Technique 23(2):709-715.
Johnson, S. 2006. The ghost map. New York: Riverhead Books. P. 219.
Kaiser, R., D. Coulombier, M. Baldari, D. Morgan, and C. Paquet. 2006. What is epidemic intelligence, and how is it being improved in Europe? Eurosurveillance 11(2), http://www.eurosurveillance.org/ew/2006/060202.asp#4 (accessed May 15, 2007).
Keystone, J. S., P. E. Kozarsky, and D. O. Freedman. 2001. Internet and computer-based resources for travel medicine practitioners. Clinical Infectious Diseases 32(5):757-765.
Kitron, U. 1998. Landscape ecology and epidemiology of vector-borne diseases: Tools for spatial analysis. Journal of Medical Entomology 35(4):435-445.
Kulldorff, M., and N. Nagarwalla. 1995. Spatial disease clusters: Detection and inference. Statistics in Medicine 14(8):799-810.
Kulldorff, M., R. Heffernan, J. Hartman, R. Assuncao, and F. Mostashari. 2005. A space-time permutation scan statistic for disease outbreak detection. PLoS Medicine 2(3):e59.
Larkin, M. 2007. Technology and public health: HealthMap tracks global diseases. Lancet Infectious Diseases 7:91.
Lescano, A. G., M. Oritz, R. Elgegren, E. Gozzer, E. Saldarriaga, I. Soriano, I. Martos, M. Negrete, and T. M. Batsel. 2003. Alerta DISAMAR: Innovative disease surveillance in Peru. American Journal of Tropical Medicine and Hygiene 69(3):S363.
Lescano, A. G., G. Salmon, and D. L. Blazes. 2007. Outbreaks investigation and control training, http://www.nmrcd.med.navy.mil/outbreak/ (accessed May 14, 2007).
Madoff, L. C. 2004. ProMED-mail: An early warning system for emerging diseases. Clinical Infectious Diseases 39(2):227-232.
Madoff, L. C., and J. P. Woodall. 2005. The Internet and the global monitoring of emerging diseases: Lessons from the first 10 years of ProMED-mail. Archives of Medical Research 36(6):724-730.
Mandl, K. D., J. M. Overhage, M. M. Wagner, W. B. Lober, P. Sebastiani, F. Mostashari, J. A. Pavlin, P. H. Gesteland, T. Treadwell, E. Koski, L. Hutwagner, D. L. Buckeridge, R. D. Aller, and S. Grannis. 2004a. Implementing syndromic surveillance: A practical guide informed by the early experience. Journal of the American Medical Informatics Association 11(2):141-150.
Mandl, K. D., B. Reis, and C. Cassa. 2004b. Measuring outbreak-detection performance by using controlled feature set simulations. Morbidity and Mortality Weekly Report 53(Suppl):130-136.
Mawudeku, A., and M. Blench. 2006. Global Public Health Intelligence Network (GPHIN), http://www.mt-archive.info/MTS-2005-Mawudeku.pdf (accessed May 8, 2007).
McNiell, W. H. 1977. Plagues and peoples. New York: Doubleday.
M’Ikanatha, N. M., D. D. Rohn, C. Robertson, C. G. Tan, J. H. Holmes, A. R. Kunselman, C. Polachek, and E. Lautenbach. 2006. Use of the Internet to enhance infectious disease surveillance and outbreak investigation. Biosecurity and Bioterrorism: Biodefense Strategy, Practice, and Science 4(3):293-300.
Morens, D. M., G. K. Folkers, and A. S. Fauci. 2004. The challenge of emerging and reemerging infectious diseases. Nature 430(6996):242-249.
Morse, S. S. 1995. Factors in the emergence of infectious diseases. Emerging Infectious Diseases 1(1):7-15, http://www.cdc.gov/ncidod/eid/vol1no1/morse.htm (accessed April 19, 2007).
Morse, S. S., B. H. Rosenberg, J. Woodall, ProMED Steering Committee Drafting Subgroup. 1996. Global monitoring of emerging diseases: Design for a demonstration program. Health Policy 38(3):135-153.
Mundana, C. C., M. Morán, M. Ortiz, E. Saldarriaga, J. Quispe, R. V. Araujo, E. Gozzer, and D. L. Blazes. 2005. Use of an electronic disease surveillance system in a remote, resource-limiting setting: Alerta DISAMAR in Peru. Abstract presented at American Society of Tropical Medicine and Hygiene Meeting, Atlanta, GA.
Mykhalovskiy, E., and L. Weir. 2006. The Global Public Health Intelligence Network and early warning outbreak detection: A Canadian contribution to global public health. Canadian Journal of Public Health 97(1):42-44.
Nsubuga, P., M. E. White, S. B. Thacker, M. A. Anderson, S. B. Blount, C. V. Broome, T. M. Chiller, V. Espitia, R. Imtiaz, D. Sosin, D. F. Stroup, R. V. Tauxe, M. Vijayaraghavan, and M. Trostle. 2006. Public health surveillance: A tool for targeting and monitoring intervention. In Disease control priorities in developing countries. 2nd ed., edited by D. T. Jamison, J. G. Breman, A. R. Measham, G. Alleyne, M. Claeson, D. B. Evans, P. Jha, A. Mills, and P. Musgrove. New York: Oxford University Press.
Olmsted, S. S., J. D. Grabenstein, A. K. Jain, W. Comerford, P. Giambo, P. Johnson, J. Mopsik, S. R. Zimmerman, and N. Lurie. 2005. Use of an electronic monitoring system for self-reporting smallpox vaccine reactions. Biosecurity and Bioterrorism: Biodefense Strategy, Practice, and Science 3(3):198-206.
Olson, K. L., M. Bonetti, M. Pagano, and K. D. Mandl. 2005 (June 21). Real time spatial cluster detection using interpoint distances among precise patient locations. BMC Medical Informatics and Decision Making 5:19.
Oxford, J. S., R. Lambkin, A. Sefton, R. Daniels, A. Elliot, R. Brown, and D. Gill. 2005. A hypothesis: The conjunction of soldiers, gas, pigs, ducks, geese and horses in northern France during the Great War provided the conditions for the emergence of the “Spanish” influenza pandemic of 1918-1919. Vaccine 23(7):940-945.
Perkins, B. A., T. Popovic, and K. Yeskey. 2002. Public health in the time of bioterrorism. Emerging Infectious Diseases 8(10):1015-1018.
Petersen, J. E. 2005. [Traveller’s medicine on the Internet]. Ugeskrift for Laeger 167(42): 3971-3973.
Reis, B. Y., and K. D. Mandl. 2003a. Integrating syndromic surveillance data across multiple locations: Effects on outbreak detection performance. Proceedings of the AMIA Symposium. Pp. 549-553.
Reis, B. Y., and K. D. Mandl. 2003b. Time series modeling for syndromic surveillance. BMC Medical Informatics and Decision Making 3(2).
Reis, B. Y., M. Pagano, and K. D. Mandl. 2003. Using temporal context to improve biosurveillance. Proceedings of the National Academy of Sciences 100(4):1961-1965.
Wagner, M. M., F. C. Tsui, J. U. Espino, V. M. Dato, D. F. Sittig, R. A. Caruana, L. F. McGinnis, D. W. Deerfield, M. J. Druzdzel, and D. B. Fridsma. 2001. The emerging science of very early detection of disease outbreaks. Journal of Public Health Management and Practice 7(6):51-59.
White House. 1996. Presidential Decision Directive, NSTC-7.
Whiteside, A., and R. Winsbury. 1996. Vancouver AIDS conference: Special report. The role of the military: To protect society—and themselves. AIDS Analysis Africa 6(4):4.
WHO (World Health Organization). 2007. Epidemic and pandemic alert and response. Disease Outbreak News, http://www.who.int/csr/don/en/ (accessed April 4, 2007).
Wilson, M. E. 1995. Travel and the emergence of infectious diseases. Emerging Infectious Diseases 1(2):39-46.
Woodall, J. 1997. Official versus unofficial outbreak reporting through the Internet. International Journal of Medical Informatics 47(1-2):31-34.
Woodall, J. P. 2001. Global surveillance of emerging diseases: The ProMED-mail perspective. Cadernos de Saúde Pública 17(Suppl):147-154.
Woodall, J., and C. H. Calisher. 2001. ProMED-mail: Background and purpose. Emerging Infectious Diseases 7(3 Suppl):563, http://www.cdc.gov/ncidod/eid/vol7no3_supp/pdf/woodall.pdf (accessed May 15, 2007).
Worldmapper. 2007. Total population, http://www.worldmapper.org/display.php?selected=2# (accessed May 15, 2007).
Zlotnik, H. 2005 (April 4). Statement to the Thirty-Eighth Session on the Commission on Population and Development, http://www.un.org/esa/population/cpd/cpd2005/Statement_HZ_open.pdf (accessed May 15, 2007).