Digital Holographic Microscopy for 3D Imaging of Complex Fluids and Biological Systems
VINOTHAN N. MANOHARAN
Harvard University
Cambridge, Massachusetts
In the past decade, three-dimensional optical imaging has emerged as a leading tool for scientific discovery in many fields, including condensed-matter physics, materials science, and biology. Confocal microscopy and related techniques provide quantitative characterizations of both the structure and dynamics of systems of many interacting microscopic components. In materials science and physics, these real-space imaging techniques have revealed in enormous detail the mechanisms of phase transitions and nonequilibrium phenomena such as glass formation—mechanisms that cannot in general be resolved using ensemble-averaging techniques such as scattering (van Blaaderen and Wiltzius, 1995; Weeks et al., 2000; Yethiraj and van Blaaderen, 2003). In addition, these techniques have led to a better quantitative understanding of many biological processes, including cell adhesion (Discher et al., 2005), mechanosensing (Ingber, 2003), and nucleoid formation in bacteria (Bates and Kleckner, 2005; Jun and Mulder, 2006).
Building up a three-dimensional image using optical microscopy requires mechanically scanning the field of view through a relatively thin sample. Thus these techniques are limited to studying processes that occur on time scales slower than the acquisition time, which is on the order of one second. An addition, confocal microscopes cost hundreds of thousands of dollars. This paper describes a promising interferometric technique, digital holographic microscopy (DHM), that is designed to overcome many of the limitations of optical microscopy and enable new experiments and new scientific discoveries.
DIGITAL HOLOGRAPHIC MICROSCOPY
Principles
DHM (Garcia-Sucerquia et al., 2006; Schnars and Juptner, 2002) is based on a technique originally outlined by Denis Gabor in 1949. A simple, in-line holographic microscope (Figure 1) consists of a laser, a camera, and an objective lens. Laser light scatters off structures in a microscopic sample, and the camera images the interference pattern between the scattered and unscattered light. The image is called a hologram.
To see how this technique can lead to a three-dimensional image, consider a coherent plane wave scattering off an idealized point particle. This particle represents anything that scatters light in the microscopic sample (e.g., a nanoparticle in solution or a subcellular structure). If we place a screen directly in front of the particle, we can see a fringe pattern that arises from interference between the scattered wave and the unscattered portion of the plane wave. This interference pattern has concentric circular fringes of varying intensity. The number and diameter of the fringes varies with the distance of the screen from the particle, and therefore, the interference pattern tells us about the location of the particle in all three dimensions.
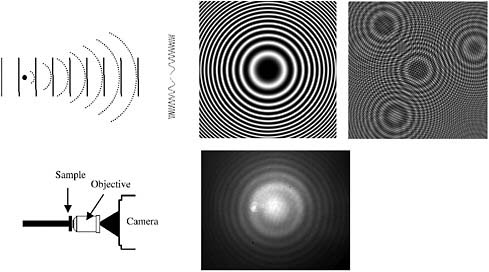
FIGURE 1 Top left: Hologram formation from a point source showing a plane reference wave interfering with a spherical scattered wave to produce (center, calculated) a Gabor zone plate pattern. Right, calculated: A set of point scatterers yields the coherent superposition of zone plates. Bottom center: Diagram of in-line holographic instrument (bottom left); image of hologram from 1 μm diameter polystyrene particle in water.
If the interference pattern is exposed onto photographic film, it produces an amplitude pattern called a Gabor zone plate. The film containing the zone plate now functions as a type of diffractive lens; if we shine a plane wave back through the developed film, it will come to a focus at a point exactly where our point particle was. If the object consists of many point particles distributed through space, then the interference pattern is simply the coherent superposition of Gabor zone plates. As Gabor first noted, when the scattering from the particles is weak, one can shine a plane wave back through the hologram (the photograph of the interference pattern) and recover an accurate, three-dimensional reconstructed image of the object.
In DHM, the hologram is captured on a digital camera, and the reconstruction is done by numerically solving the Fresnel-Kirchhoff diffraction equation—in effect, digitally “shining light” back through the hologram. Today nearly all holography is digital. Because the hologram of a set of point sources is mathematically equivalent to the Fourier convolution of zone plate patterns, one can use Fourier-transform methods for the reconstruction (Kreis, 2002).
Holography, therefore, enables us to obtain a volume rendering of a three-dimensional object or particle distribution from a single two-dimensional image. This means DHM is, in principle, a fast, real-space, three-dimensional imaging technique. Compared to other 3D techniques, such as tomography (Xu et al., 2001) and confocal microscopy (Weeks et al., 2000), DHM is potentially 3 orders of magnitude faster in acquisition time.
Moreover, DHM, especially in the in-line configuration that requires only a laser, an objective, and a camera, is simple and cheap to build. The major cost is the huge increase in processing time required to reconstruct the 3D image. DHM is not a new technique, but until recently its applications have been limited because of the high computational cost of processing holograms. The availability of fast computers for image processing, inexpensive semiconductor lasers, and high-speed CMOS cameras—all made possible by advances in semiconductor technology—have made it possible to build holographic microscopes cheaply and easily in the laboratory.
Quantitative Holographic Microscopy
Because the particles imaged in the hologram, which form the structure of the sample, can be tracked through space and time with high spatial precision, DHM can be used as a quantitative imaging technique. Like many other particle-tracking techniques (Crocker and Grier, 1996), there is some confusion over the word “resolution.” In holography, as in all optical techniques, the spatial resolution is limited by diffraction; at best, one can resolve two point sources about a wavelength apart, but not much closer than that. However, it is possible to resolve the center of brightness of a single spherical particle to a precision on the order of tens of nanometers, well below the diffraction limit.
For example, using a DHM built with a diode laser and a high-speed CMOS camera, my research group has captured holograms of a 1 μm polystyrene sphere diffusing in water as a function of time. By reconstructing these images and tracking the centroid of the particle, we can measure the diffusion coefficient in all three dimensions to within 5 percent of the value expected from the Stokes-Einstein relation. DHM can accurately measure particle dynamics in real space, in three dimensions, with spatial precision of about 10 nanometers (nm) and temporal resolution of milliseconds. Even smaller time scales (tens of microseconds) are possible with faster cameras.
HOLOGRAPHIC MICROSCOPY FOR STUDYING SELF-ASSEMBLY IN COMPLEX FLUIDS
We use DHM primarily for studying the self-assembly of complex fluids, and in particular colloidal and nanoparticle suspensions; the particles in these systems have diameters of 10 to 1,000 nm. Interactions among these particles can drive them to self-assemble into ordered structures at equilibrium, such as colloidal crystals, which can serve as the basis for advanced functional materials like photonic crystals (Vlasov et al., 2001). Thus, at least in principle, self-assembly represents a cheap and easy way to fabricate advanced materials (Dinsmore et al., 1998, 2002; Klein et al., 2005; Manoharan and Pine, 2004). However, we do not yet understand the dynamic processes involved in self-assembly. DHM is one of the few experimental tools that can resolve the particle positions and dynamics in these small, rapidly changing, isolated systems.
A small self-assembling system consisting of colloidal particles on the surface of a spherical oil droplet is shown in Figure 2 (McGorty et al., 2008). Altogether, there are 10 polymer particles, each with a diameter of about 800 nm, that interact on the surface of the droplet to form an ordered polyhedron. Such small self-assembled structures, which are common in industrial fluids and personal care products, are also useful as building blocks for new materials. But, because of droplet rotation and particle motion, such systems are nearly impossible to probe in three dimensions with confocal microscopy.
With DHM, however, it is possible to capture the full three-dimensional positions of all of the particles at a given time in one hologram (Figure 2). We gather these fully three-dimensional data sets at a rate of 30 per second. Because the system is at equilibrium, we can use Boltzmann statistics to extract parameters, such as the potential of mean force (also shown in the figure), which is the interparticle pair potential averaged over the configurations of all other particles in the system.
The advantage of this technique is that it gives us (in theory) all the information about the system at equilibrium. With each hologram we obtain a 3D snapshot of the system at a particular spot in its configuration space. The statistics of these configurations can then be used to derive the interactions and energetics through
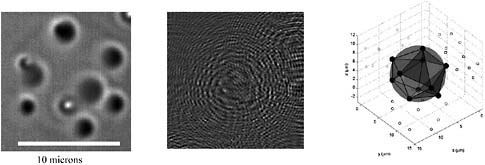
FIGURE 2 Left: Bright field microscope image of 10 particles at the interface of an emulsion droplet (index matched). Center: Hologram of same droplet taken with our DHM instrument. Right: Reconstruction of hologram showing positions of particles on the surface of the droplet and projections of positions onto coordinate planes. Reconstruction is drawn to scale. Source: McGorty et al., 2008.
the Boltzmann distribution (Crocker and Grier, 1994; Lee and Grier, 2007). These measurements are beginning to reveal the mechanism of how these interactions lead to the ordered structure seen on the surface of the droplet.
CHALLENGES AND PROSPECTS
There are still some challenges to using holography as a general-purpose 3D imaging technique. The most severe drawback is the enormous processing power, memory, and storage capacity required for computing holographic reconstructions. Today, with fast Fourier-transform algorithms, we can reconstruct a hologram for a single z-section within a few tenths of a second on a common personal computer. But we generally work with thousands of holograms taken at different times, and for each hologram we reconstruct a volume that contains about a thousand different z-sections. Thus processing a typical time series of holograms takes several days on a personal workstation, and for each 1 gigabyte holographic “movie,” we can generate nearly a terabyte of volumetric reconstructions. In the meantime, although work remains to be done before DHM can replace confocal microscopy as a general tool, it can be used to probe specific systems and important scientific questions.
One intriguing possibility is biological imaging. Several studies have demonstrated that holographic microscopy can be used to image subcellular structures in three dimensions in near-real time (Choi et al., 2007; Marquet et al., 2005; Xu et al., 2001). Holographic microscopes could greatly reduce costs and sample preparation times over confocal microscopy. In fact, the most expensive part of a DHM is the computer used to reconstruct the images. Therefore, it becomes possible to use a microscope as an “add-on” to an existing biological apparatus,
such as a cell-culture chamber, rather than as a central facility. With this goal in mind, we have started to build inexpensive holographic microscopes from diode lasers and consumer-grade digital cameras. Such instruments, which can be built for less than $1,000, might be useful for studying traction forces (Dembo et al., 1996) and cell rheology (Bursac et al., 2005; Hoffman et al., 2006), as well as for providing a more physical understanding of certain pathologies, such as sickle cell anemia (Chien, 1987).
ACKNOWLEDGMENTS
This work was supported by CAREER award from the National Science Foundation.
REFERENCES
Bates, D., and N. Kleckner. 2005. Chromosome and replisome dynamics in e. coli: loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell 121(6): 899–911.
Bursac, P., G. Lenormand, B. Fabry, M. Oliver, D.A. Weitz, V. Viasnoff, J.P. Butler, and J.J. Fredberg. 2005. Cytoskeletal remodelling and slow dynamics in the living cell. Nature Materials 4(7): 557–561.
Chien, S. 1987. Red cell deformability and its relevance to blood flow. Annual Review of Physiology 49(1): 177–192.
Choi, W., C. Fang-Yen, K. Badizadegan, S. Oh, N. Lue, R.R. Dasari, and M.S. Feld. 2007. Tomographic phase microscopy. Nature Methods 4(9): 717–719.
Crocker, J.C., and D.G. Grier. 1994. Microscopic measurement of the pair interaction potential of charge-stabilized colloid. Physical Review Letters 73(2): 352–355.
Crocker, J.C., and D.G. Grier. 1996. Methods of digital video microscopy for colloidal studies. Journal of Colloid and Interface Science 179(1): 298–310.
Dembo, M., T. Oliver, A. Ishihara, and K. Jacobson. 1996. Imaging the traction stresses exerted by locomoting cells with the elastic substratum method. Biophysical Journal 70(4): 2008–2022.
Dinsmore, A.D., J.C. Crocker, and A.G. Yodh. 1998. Self-assembly of colloidal crystals. Current Opinion in Colloid and Interface Science 3(1): 5–11.
Dinsmore, A.D., M.F. Hsu, M.G. Nikolaides, M. Marquez, A.R. Bausch, and D.A. Weitz. 2002. Colloidosomes: self-assembled, selectively permeable capsules composed of colloidal particles. Science 298(5595): 1006–1009.
Discher, D.E., P. Janmey, and Y.L. Wang. 2005. Tissue cells feel and respond to the stiffness of their substrate. Science 310(5751): 1139–1143.
Gabor, D. 1949. Microscopy by reconstructed wave-fronts. Proceedings of the Royal Society A Mathematical, Physical and Engineering Sciences 197(1051): 454–487.
Garcia-Sucerquia, J., W. Xu, S.K. Jericho, P. Klages, M.H. Jericho, and H.J. Kreuzer. 2006. Digital in-line holographic microscopy. Applied Optics 45(5): 836–850.
Hoffman, B.D., G. Massiera, K.M. Van Citters, and J.C. Crocker. 2006. The consensus mechanics of cultured mammalian cells. Proceedings of the National Academy of Sciences 103(27): 10259–10264.
Ingber, D.E. 2003. Mechanosensation through integrins: cells act locally but think globally. Proceedings of the National Academy of Sciences 100(4): 1472–1474.
Jun, S., and B. Mulder. 2006. Entropy-driven spatial organization of highly confined polymers: lessons for the bacterial chromosome. Proceedings of the National Academy of Sciences 103(33): 12388–12393.
Klein, S.M., V.N. Manoharan, D.J. Pine, and F.F. Lange. 2005. Synthesis of spherical polymer and titania photonic crystallites. Langmuir 21(15): 6669–6674.
Kreis, T.M. 2002. Frequency analysis of digital holography with reconstruction by convolution. Optical Engineering 41(8): 1829–1839.
Lee, S.H., and D.G. Grier. 2007. Holographic microscopy of holographically trapped three-dimensional structures. Optics Express 15(4): 1505–1512.
Manoharan, V.N., and D.J. Pine. 2004. Building materials by packing spheres. MRS Bulletin 29(2): 91–95.
Marquet, P., B. Rappaz, P.J. Magistretti, E. Cuche, Y. Emery, T. Colomb, and C. Depeursinge. 2005. Digital holographic microscopy: a noninvasive contrast imaging technique allowing quantitative visualization of living cells with subwavelength axial accuracy. Optics Letters 30(5): 468–470.
McGorty, R., J. Fung, D. Kaz, S. Ahn, and V.N. Manoharan. 2008. Measuring Dynamics and Interactions of Colloidal Particles with Digital Holographic Microscopy. In Digital Holography and Three-Dimensional Imaging, OSA Technical Digest. Available online at http://www.opticsinfobase.org/DirectPDFAccess/30DA92AA-BDB9-137E-C8A9FB62AAFF878B_156213.pdf?da=1&id=156213&seq=0.
Schnars, U., and W.P.O. Juptner. 2002. Digital recording and numerical reconstruction of holograms. Measurement Science & Technology 13(9): R85–R101.
van Blaaderen, A., and P. Wiltzius. 1995. Real-space structure of colloidal hard-sphere glasses. Science 270(5239): 1177–1179.
Vlasov, Y.A., X.Z. Bo, J.C. Sturm, and D.J. Norris. 2001. On-chip natural assembly of silicon photonic bandgap crystals. Nature 414(6861): 289–293.
Weeks, E.R., J.C. Crocker, A.C. Levitt, A. Schofield, and D.A. Weitz. 2000. Three-dimensional direct imaging of structural relaxation near the colloidal glass transition. Science 287(5453): 627–631.
Xu, W.B., M.H. Jericho, I.A. Meinertzhagen, and H.J. Kreuzer. 2001. Digital in-line holography for biological applications. Proceedings of the National Academy of Sciences 98(20): 11301–11305.
Yethiraj, A., and A. van Blaaderen. 2003. A colloidal model system with an interaction tunable from hard sphere to soft and dipolar. Nature 421(6922): 513–517.








