8
Fostering the Global Dimension of the Health Data Trust
INTRODUCTION
The ability to draw broadly from anywhere across the globe to provide relevant insights for health and healthcare improvement is a long-term goal for the learning health system. Meanwhile, the ability to learn from the experiences of other countries and to apply health information technology (HIT) for biosurveillance can actively facilitate progress toward this and other goals. This chapter reviews several activities relevant to exploring the global dimension of the digital infrastructure for a learning health system.
In his paper, Brendan Delaney from Kings College London describes the TRANSFoRm project. TRANSFoRm, a European Union (EU) effort to develop a learning health system driven with HIT, has been designed based on carefully chosen clinical use cases and is aimed at improving patient safety as well as supporting and accelerating clinical research. Dr. Delaney outlines several of the challenges that have arisen such as system interoperability, a need for advanced functionalities, and the support of knowledge translation. He also describes several techniques being employed to address these challenges, including clinical research information models, service-based approaches to semantic interoperability and data standards, detailed clinical data element representations built on archetypes, and an effort to prioritize electronic health record (EHR) and workflow integration in the development of clinical decision support systems that are designed to capture and present fine-grained clinical diagnostic cues.
Drawing from his involvement with SHARE, an EU-funded project to define the path toward greater implementation of grid computing ap-
proaches to health, Tony Solomonides, from the University of the West of England, discusses his current work to automate policy and regulatory compliance to allow health information sharing. He describes the implementation of attribute-based access controls to ensure enforcement of privacy obligations which—due to variations in their interpretation between EU countries—require a logic-based computed approach.
HIT holds great promise to increase quality and improve patient safety in developing and transitional countries. Harvard University’s Ashish Jha describes how a dearth of reliable information has impeded efforts to better understand and design solutions to higher rates of adverse event–associated morbidity in developing countries, as well as obtain an accurate calculation of global disease burden. Dr. Jha describes an effort by the World Health Organization to maximize the impact of HIT in resource-poor settings through the development of a minimum dataset that would allow for systematic data collection to address safety issues.
David Buckeridge and John Brownstein from McGill University describe how HIT is enabling dramatic changes in domestic and international infectious disease surveillance. Detailing how the digital infrastructure can enhance existing systems through the use of automation and decision support, the authors also address novel approaches to surveillance enabled by recent informatics innovations. Using the DiSTRIBuTE project as an example of innovations in syndromic surveillance that drastically improve coverage and speed, they call for a renewed science of disease surveillance that embraces information technology as well as the potentially disruptive changes it brings to improve disease control.
TRANSFoRm:
TRANSLATIONAL MEDICINE AND PATIENT SAFETY IN EUROPE
Brendan Delaney, M.D.
King’s College London
The underlying concept of TRANSFoRm is to develop a “rapid learning healthcare system” driven by advanced computational infrastructure that can improve both patient safety and the conduct and volume of clinical research in Europe.
The European Union (EU) policy framework for information society and media, identifies e-health as one of the principal areas where advances in information and communications technology (ICT) can create better quality of life for Europe’s citizens (Europe’s Information Society, 2009). ICT has important roles in communication, decision making, monitoring, and learning in the healthcare setting. TRANSFoRm recognizes the need
to advance the underpinning information and computer science to address these issues in a European and international context.
The Challenge of Interoperability
Providing interoperability between different clinical systems (which span national boundaries) and integrating those systems with the research enterprise lies at the heart of the eHealth Action Plan (Iakovidis and Purcarea, 2008). In both domains fragmentation of records and proprietary systems that do not adhere to uniform standards are as much of a challenge as the legal and ethical issues that complicate access to clinical data for researchers (Delaney, 2008). However, significant advances in international standards and in computational technology to support interoperability offer a way to overcome these challenges. Furthermore, advances in the understanding of clinical judgment and decision making—as well as the ways of supporting them via ICT—can inform the design of more “intelligent”electronic health record (EHR) systems.
Interoperability of data is underpinned by shared concepts and a common terminology (or at least an agreed and maintained mapping between terminologies). In research, interoperability of concepts between domains is promoted by the Biomedical Research Integrated Domain Group (BRIDG) Model (Fridsma et al., 2008). In primary care, the Primary Care Research Object Model defines the necessary domain-specific data classes, mapped to BRIDG (Speedie et al., 2008). In addition to terminologies, the system needs to enable multilanguage representations of the clinical terms, which is particularly important from an EU perspective.
However, simply providing a mechanism for the high-level interoperability of data will not provide sufficient functionality for a learning health system. System integration and shared detailed clinical data representations are also required. The system needs to have a common business model with a shared model of processes driven by a suite of open source middleware. Further, the integration of systems requires a much deeper level of interoperability than simple “diagnosis.” Although SNOMED-CT has an underlying classification and allows for the concatenation of terms as well as representing diagnostic concepts such as clinical signs, it is probably not rich enough to represent all the symptoms and signs required for a diagnosis. Furthermore, these concepts need to be linked in an ontology rather than just a classification.
Building a Learning Health System
The single richest source of routine healthcare data lies within the records of Europe’s general practitioners. Primary care providers are re-
sponsible for first contact, continuing, and generalist care of the entire population from birth to death (Schade et al., 2006). Any project that aims to comprehensively support the integration of clinical and research data should begin with primary care. In addition, even in countries where general practitioners do not fulfill a “gatekeeper” function—controlling access to specialist services—the quality of initial diagnosis at the primary care level determines much of the future course for an individual patient. In order to support patient safety in both clinical and research settings, significant ICT challenges need to be overcome in the areas of interoperability, common standards for data integration, data presentation, recording, scalability, and security (Ohmann and Kuchinke, 2009).
To explore these issues in more depth, it is useful to consider a list of requirements for a learning health system:
- Supports complex queries of existing data, distributed and with support for various mapped terminologies.
- Supports real-time recruitment of subjects with workflow-integrated prompts based on reason for encounter or any other data item within the clinical encounter.
- Supports real-time prompts for data or sample collection based on data items within the clinical encounter.
- Supports jointly controlled data entry into research and clinical records.
- Supports real-time diagnostic and therapeutic decision support.
- Supports all relevant requirements of data privacy, consent, and security.
- Supports full audit and provenance of data.
To support this level of functionality a sharing of concepts at the very deepest level is required. The international standard CEN/ISO 13606 supports the use of archetypes (Kalra et al., 2005). Archetypes are computable expressions of a domain content model in the form of structured constraint statements based on a reference information model. They are often encapsulated together in templates, sit between lower level knowledge resources and production systems, and are independent of the interface and system. The latter is essential to the development of a sustainable business model whereby core shared work on archetypes can be deployed via a variety of commercial EHR systems.
Efficient support of knowledge translation is the final piece in the jigsaw. While decision support systems for management, quality improvement, and prescribing have all been shown to be effective, no system for diagnostic decision support has been positively evaluated or widely deployed (Garg et al., 2005). The principal reason for this is the failure of clinicians
to use the systems routinely. Not only do they not integrate seamlessly with the EHR—for the technical reasons described above—but they have been developed without an understanding of the cognitive workflow involved in diagnosis. Much recent work in the field of medical decision making indicates that there may be specific points within the diagnostic process where decision support, in the form of alerts or prompts, may be effective. Accurate diagnosis has been shown to be related to the acquisition and interpretation of critical clinical cues. This process should be amenable to support by a well-specified ontology of diagnostic cues (Kostopoulou et al., 2008). In order for this to be achieved, it is necessary to provide an EHR interface that readily supports the capture and presentation of fine-grained clinical diagnostic cues. Given that “failure to diagnose promptly”is the single most common cause of litigation against primary care physicians, detailed justification of a diagnosis—richly recorded and linked to a knowledge base—will be one means by which clinicians may reduce the risk of litigation while improving patient care (Singh et al., 2007).
The TRANSFoRm Project
International cooperation in this area is essential. Working with and extending international standards for the representation of data and machine-readable clinical trial protocols, archetypes, and terminology services require international consensus and models of shared ownership. In addition, the market within which EHR systems are developed needs to be opened up to allow for widespread adoption of innovative user interfaces, decision support, terminology, and archetype services, and the export and linkage of data. The restriction of access to EHR data and systems is anti-competitive and restricts innovation in this field.
TRANSFoRm (Figure 8-1) brings together a highly multidisciplinary consortium where three carefully chosen clinical “use cases” will drive, evaluate, and validate the approach to the ICT challenges. The project will build on existing international work in clinical trial information models (BRIDG and the Primary Care Research Object Model), service-based approaches to semantic interoperability and data standards (ISO11179 and controlled vocabulary), data discovery, machine learning, and EHRs based on open standards (CEN/ISO 13606). We will extend this work to interact with individual EHR systems as well as operate within the consultation itself, providing diagnostic support as well as support for the identification and follow-up of subjects for research. The approach to system design will be modular and standards based—providing services via a distributed architecture—and will be tightly linked with the user community. Four years of development and testing will end with a fifth year dedicated to summative validation of the project deliverables in the primary care setting.
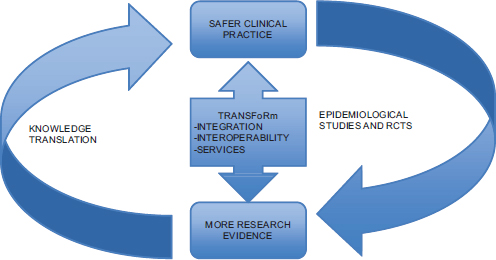
FIGURE 8-1 TRANSFoRm and the learning health system
HEALTHGRIDS, THE SHARE PROJECT, AND BEYOND
Tony Solomonides
University of the West of England
Grid computing was introduced in the late 1990s to serve as a medium of scientific collaboration and as a more immediate means of high-performance computing (Foster and Kesselman, 2004). If the Internet is an apparently inexhaustible information medium, the grid would also add rapid computation, large-scale data storage, and flexible collaboration by harnessing the power of large numbers of computers. As a computational paradigm, the grid was adopted for use in scientific fields—such as particle physics, astronomy, and bioinformatics—in which large volumes of data, very rapid processing, or both, are necessary.
The complementary idea of e-science arose from the observation that a scientist often has to juggle experiments, data collection, data processing, analysis of results, and their iteration and refinement. There is a need for intelligent conduit of information between these processes. Why not facilitate this through an informatic infrastructure that allows the scientist to pipeline activities in some way, leaving her free to concentrate on the science? If the work is being undertaken together with other scientists, this infrastructure should also support their collaboration but not expose their individual or joint efforts to anyone outside the specified group of collaborators.
Grids and Clouds in Health Care
There have also been several ambitious medical and healthcare applications of grids. While these initial exemplars have been mainly restricted to the research domain, there is a great deal of interest in real-world applications. However, there is some tension between the spirit of the grid paradigm and the requirements of healthcare applications. The grid maximizes its flexibility and minimizes overheads by requesting that computations be performed, and data stored/replicated, at the most appropriate node in the network. On the other hand, a hospital or other healthcare organization is required to maintain control of its confidential patient data and to remain accountable for its use at all times. The very basis of grid computing therefore appears to threaten certain inviolable principles: the confidentiality of medical data, the accountability of healthcare professionals, and the precise attribution of “duty of care.”
Cloud computing is a more recent but related innovation. Like the grid, it arises from concepts and forces that were already present in the field, not least in the world of commercial computing. Precursors include the ideas of “application service provision” and “virtualization.” Indeed, early adaptations of concepts from grid computing included the notion of utility computing—computing power distributed as if it were a “domestic” utility like gas of electricity. The advantage to a business that outsources its information systems to a cloud provider is that it need not own the infrastructure of servers and communications nor concern itself with maintaining the applications.
The current convergence of utility computing with social networking applications has led to several serious proposals to use clouds for patient-, or more accurately, carer-managed electronic health records (EHRs): commercial examples include Microsoft’s HealthVault and Google Health, while in the United Kingdom there is debate on extending the use of HealthSpace along such lines. Indeed, the idea that personal EHRs could be “banked” originated with Dr. Bill Dodd in 1997 (Dodd, 1997). The opportunity to mine such records to the advantage of public health has also been noted (Bonander and Gates, 2010).
Healthgrids arose from the observation that healthcare and biomedical research share many of the characteristics of e-science. Consequently, many areas of biomedical research—medical imaging and image processing, modeling the human body, pharmaceutical research and development, epidemiological studies, genomic research, and personalized medicine—are expected to benefit from healthgrid technology. To use a familiar and successful example, consider a patient in a breast cancer screening program. If a mammogram gives cause for concern, it may be necessary to conduct further investigation or to seek a second opinion. There is already a powerful array
of technological support for this, from image standardization software to computer-aided detection. The possibility of remote second opinion is also considered valuable if it does not take up too much time. If the patient is referred, the oncologist wants to know the history as succinctly as possible in order to review the diagnosis and begin with assessment and staging. If the patient needs to undergo surgery, the images from the diagnostic stage can be used in planning. In other cancers, radiotherapy planning may be assisted by review of imaging (Warren et al., 2007).
A powerful influence over the direction of these early projects was the Bioinfomed study which established a now familiar picture of the correspondences between biosocial organization (molecule–cell–organ–individual–community), pathologies and disciplines with different kinds of informatics (molecular modeling–imaging of cells and organs–electronic patient records–public health informatics) (Martin-Sanchez et al., 2004). It challenged the community to bring together information at these different levels into a coherent model. One of its most obvious successors is the Virtual Physiological Human, a program that seeks to provide a framework for the integration of different partial models of the human body, on different scales, toward an aggregate systemic study of human physiology.
HealthGrid and SHARE
HealthGrid was an EU-inspired initiative to support projects in the use of grid technology in health care and biomedical research. Incorporated as a not-for-profit organization in France, this collaboration edited a white paper setting out for senior decision makers the concept, benefits, and opportunities offered by healthgrids (Vincent et al., 2005). Starting from these conclusions, the EU funded the SHARE project aimed at identifying the important milestones toward wide deployment and adoption of healthgrids in Europe, perhaps as part of an action plan for a “European e-Health Area” (SHARE Collaboration, 2008). The project had to assess the status quo and set targets; identify key gaps, barriers, and opportunities; establish short- and long-term objectives; propose key developments; and suggest the actors needed to achieve the vision. The road map had to encompass issues regarding networks; infrastructure deployment; “middleware”; services to end users; standards; security; ethical, legal, and regulatory developments; social adjustments; and economic investments.
A draft road map was filtered through a number of “use cases” including drug discovery, large-scale public health emergency, imaging-based screening, and management of chronic conditions. The requirements arising from these different case studies led to differentiation between the development of (1) data, (2) computational, and (3) collaboration healthgrids. Indeed, the third category crystallized in the course of the project. The
ultimate goal of a “knowledge grid” was then seen to emerge from the interaction of these three subparadigms, rather than to be an enhancement of the data grid, as had previously been thought.
Ethical, legal, social, and economic issues assumed increasing importance in the course of the project. The project mapped the legal and ethical landscape, identifying barriers to the wide adoption of healthgrids. Aspects of the law and emphasis on ethical requirements were initially considered to be inert constraints but were subsequently treated as parallel dynamic developments capable of being influenced by policy. These were therefore included in the road maps as areas in which fresh thinking and strategy were necessary. A project since undertaken at University of the West of England, Bristol, has demonstrated that it is possible for technology to incorporate goals such as regulatory compliance even in the face of potentially contradictory demands from different frameworks.
In relation to health care, SHARE identified evidence-based practice as the core requirement. As such, much of the work is underlain by assumptions about the dynamic nature of the evidence base, the need for biomedical advances to be translated into medicine, and for gold standard evidence to be interpreted in operational terms. Arguably, it paid less attention to the business of health care, including “internal markets” and commissioning (as in the United Kingdom) or actual markets (as in the United States). For example, the possibility of patients owning their data in real rather than in moral terms was considered but not fully explored. Developments in healthcare systems—including the halting progress of the English National Health Service National Programme for IT—have led governments to consider the role of cloud computing for the management of electronic patient records. This is regarded as a positive development that should help close the gap between healthgrids (for science and knowledge management) and clouds (for management, compliance, and business issues).
Technology and Regulatory Compliance
It has already been observed that the grid paradigm is in some ways at odds with the requirements of healthcare organizations. Although it featured significantly in subsequent research, security was not a top priority in its initial development. However, the complexity of medical data, the risk of disclosure through metadata, and the granularity of confidentiality are not readily accommodated in a raw grid environment. Healthgrids would have to take account of these constraints if they were ever to succeed in biomedical research or healthcare. Yet, all advantage would be lost if the very efficiency of grid computing was undermined by a constant need for human regulatory intervention.
The situation is somewhat reminiscent of the history of the motor car.
When the first motorized carriage was introduced in England in the mid-1890s, it was a legal requirement that a man walk ahead of any motorized vehicle with a red flag to warn pedestrians and to ensure that its speed did not exceed 4 mph.1 It would be absurd to impose a restriction of that nature on healthgrids. The very idea behind the concept was to make sharing and exchange of data and workflows as smooth and uninterrupted as possible. Our goal in subsequent research was to show that technology could at least meet legal and ethical regulatory frameworks halfway. In doing so, technological innovation as well as ethical and legal policies would be framed in ways that acknowledged each other’s legitimate concerns. Along with proposals for the mutual education of technologists and policy makers, this project was intended to be a demonstrator not only of technology applied to regulation, but of technology developed in the light of a sometimes uncertain and occasionally self-contradictory regulatory framework.
In the European Union, many areas of activity are controlled by what are known as “directives.” For example, the European Working Time Directive restricts the number of working hours for different kinds of work. However, European directives are not legislation. Each directive has to be “transposed” as national legislation separately by each member state. Consequently, there is no guarantee of consistency. In our case, the relevant directive is 95/46/EC Data Protection Directive (European Parliament and Council of the European Union, 2010). The definitions of relevant terms (e.g., “personal data”) and restrictions on data disclosure vary from country to country, even though all legislation is supposed to correspond to 95/46/EC. At the heart of the project reported here, therefore, is an assumption that text law is too complex to be interpreted by nonlegal expert users of healthgrids—whether they are biomedical researchers, clinicians, or technologists. Thus, we propose a twin-track approach: on one hand, the system may offer advice and decision support; on the other, it can ensure enforcement of privacy obligations at the process level (Figure 8-2).
At its most abstract, the initial question was this: given some legislation that has been translated into some sort of declarative framework, could we take that and map it to a deontic logic of permissions and obligations. In other words, can we develop an operational logic that could function at the infrastructure level? This begs the question: What sort of declarative framework would be suitable to encode legislation? The problem factors in a variety of ways. One of these is to distinguish between actionable advice and operational permissions/obligations. More importantly, the problem also factors into “preconditions for access to the data” and “postconditions for the treatment of the data.” Finally, since much compliance checking is
___________________
1 See http://www.datchethistory.org.uk/Link%20Articles/Ellis/evelyn_ellis.htm (accessed September 10, 2010).
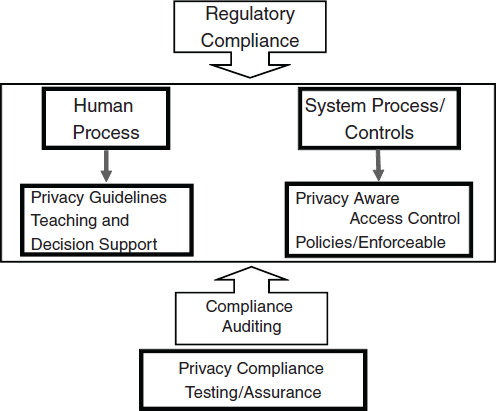
FIGURE 8-2 Proposed regulatory framework for the HealthGrid.
done through audit, we need to determine what to document, and how, in order to provide evidence for audit.
A Proposed Ontology for Data Sharing
An approach through ontology allows us to (1) provide a semantic map of the directive and its “transposition” into UK, French, and Italian legislation; and (2) use the so-called Semantic Web Rule Language (SWRL) to reason with the ontology.
Figure 8-3 gives a diagrammatic representation of the Protégé ontology for rules on data sharing. At its center is an event of proposed DataSharing, which relates to certain data to be shared (SharedData) whose Privacy Status (Anonymized, Encrypted, or Raw) is also known. The DataSharing has a Sender and a Receiver, both of which, along with the SharedData, belong to a MemberState. The DataSharing has a SharingPurpose. Based on this information we can determine the ConsentNecessity (Necessary or Unnecessary), ConsentSpecificity (Specific or Broad), ConsentExplicitness
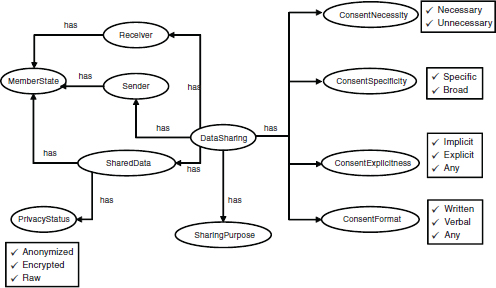
FIGURE 8-3 Ontology model of data-sharing contexts and requirements.
(Implicit, Explicit, or Any—that is, either or perhaps not even known), and the ConsentFormat (Written, Verbal, or Any).
Taking as our example a permissive clause, we consider the preconditions under which it is applicable and postconditions in the form of obligations or constraints on any subsequent processing of the data. The condition on the user(s), the data, and the purpose of any proposed sharing of the data are given in the Web Ontology Language (World Wide Web Consortium, 2009). SWRL is used to translate this knowledge into an if-then action rule, whose consequent involves an Action (e.g., Allow) and the imposition of certain further Obligation(s) on how the data should be processed once the permission has been enacted (World Wide Web Consortium, 2004).
A typical scenario may be the following: Patient Emma’s mammogram series gives Dr. House some cause for concern; he believes that the mammogram includes certain features that Dr. Casa in Italy has reliably diagnosed with great accuracy in the past. Emma has provided consent for the mammograms to be taken and processed for the purpose of “breast cancer diagnosis and treatment.” Dr. House’s purpose in sharing the data with Dr. Casa is “to obtain second opinion on treatment options” which is compatible with the purpose for which Emma gave consent. The mammogram has been stripped of all obviously identifying information, but it could be traced back to Emma through secondary attributes and information about where and by whom she was treated. Nevertheless, for the strict clinical purpose
for which the sharing is proposed, transmission of the mammogram to Dr. Casa is approved (provided he will destroy his electronic copy once he has completed his diagnosis). Thus, if Dr. Casa’s insurance requires him to keep a record for his own protection, or if he wishes to use the mammogram in a text book, he must request further permission to do so. The SWRL representation of this example is shown in Figure 8-4.
Our intention was to translate our SWRL rules into an actionable logic. The choice for this is the eXtensible Access Control Markup Language (XACML) (OASIS, 2005). In XACML, a Policy is made up of Rules which may be combined through a Rule Combining Algorithm (it is also possible to have Policy Sets with Policy Combining Algorithm). A Policy may impose a certain Obligation as part of its response. A Rule has a Target and an Effect (e.g., allow or deny). A Target (i.e., the object of the Rule) includes a Subject (to whom the response is directed), an allowed or disallowed Action, a Resource to which the Action applies, and a Purpose for which the Action would be taken. Key structural elements in an XACML implementation are the Policy Decision Point and the Policy Enforcement Point. In our case, the Policy Information Point here has been implemented as our Semantic Web Knowledge Base and the Context Handler.
Figure 8-5 depicts this model and the numbered arrows indicate the sequential data flow that implements the rule we gave above. In the event that a whole set of data is to be shared, the same process takes place, except that the Context Handler classifies the data into sets with similar pre- and postconditions. The Context Handler now communicates directly with the Policy Enforcement Point to provide information, although the decision, as ever, is issued by the Policy Decision Point.

FIGURE 8-4 Semantic Web Rule Language representation of a data-sharing event.
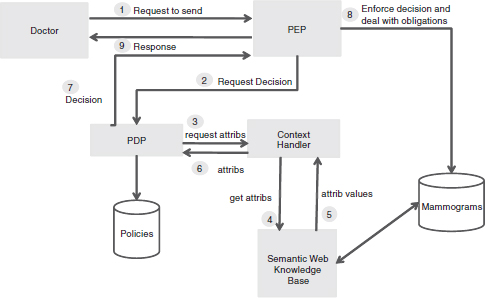
FIGURE 8-5 Extended XACML access control model/data flow.
Conclusion
These examples show how we may model contexts of medical data sharing by means of ontology, reason about which privacy requirements should be assigned to them, extend the ontology to allow the specification of adequate attribute-based access control policies, and map the semantic web policies to XACML to prove enforceability. The technological solution outlined above can handle the ambiguity of rules in the face of different interpretations of the same directive. The combining algorithm may be set to be conservative or liberal, maximal or minimal; in neither case does it violate any principles. In some circumstances it may not be able to reach an unambiguous decision, referring the user to authority.
Hopefully, this model points to a solution not only to the problem of automating compliance checks and speeding up the process of sharing medical data, but also to the issue of provenance management—that is, maintaining a metarecord with the data that provides details of where it came from, how it was constructed, what processes it has undergone since, and so on. This facilitates research through secondary use as well as the legal process of audit of compliance.
A GLOBAL PERSPECTIVE ON THE IMPORTANCE OF
SYSTEMATIC DATA TO DRIVE IMPROVEMENTS IN CARE
Ashish K. Jha, M.D., M.P.H.
Harvard School of Public Health
There is broad consensus that improving patient safety is a critical component of advancing the health and well-being of citizens across the globe. Policy makers and clinicians increasingly view health information technologies (HITs)—and the data that underlie these systems—as a tool to drive quality improvement and improve patient safety. To date, the vast majority of global health efforts have focused on promoting access to care in developing and transitional countries. These efforts have further focused on specific conditions commonly viewed as the major global killers: HIV/AIDS, tuberculosis, and malaria.
Despite the successes realized by many of these initiatives, patient safety is an area of significant concern that warrants heightened attention among policy makers. Surprisingly, we know little about the safety of care delivered to patients and the magnitude to which care may cause harm. The available evidence indicates that unsafe care is a major cause of morbidity, mortality, and years of life lost, also carrying significant financial implications on health systems and society. Yet, due to the lack of systematic data sources, there is a dearth of data to inform actionable strategies aimed at improving the safety of care.
In this context, HIT may play a meaningful role. While the use of HIT systems to improve the safety and effectiveness of care delivered has received considerable attention in developed nations, the global debate on how HIT systems may be used to improve care in developing and transitional nations is in its infancy. The majority of key data needed to help policy makers and decision makers prioritize funding and allocate resources simply do not exist. Developing even the most basic form of information infrastructure is critical to thoughtfully push forward the policy debate. To better understand how HIT may be most effective, and to identify the best areas for intervention, more research is needed on the safety of care delivered in developing, transitional, and developed nations.
WHO World Alliance for Patient Safety
The World Health Organization (WHO) World Alliance for Patient Safety Working Group was charged with identifying global priorities for patient safety research. The group undertook two major initiatives: a report on the state of evidence on patient safety and calculating the global burden of unsafe care
Report on the State of Evidence on Patient Safety
The report, Summary of the Evidence on Patient Safety: Implications for Research, provides the most comprehensive picture of adverse events in health care (Jha, 2008). The report aims to not only describe the scope of challenges facing policy makers around patient safety, but also to provide recommendations and priorities for research. Members of the working group consisted of experts with multidisciplinary expertise in epidemiology, qualitative methods, and human factors and were from developing, transitional, and developed nations in all seven WHO regions.
Initially, the group identified the types of adverse events in health care and their causes. From these efforts, a list 23 major harms and their underlying causes was created (Table 8-1). Although these topics are not comprehensive of all epidemiological and clinical metrics, they are among the most important. The 23 patient safety topics were then categorized
TABLE 8-1 World Alliance for Patient Safety List of Common Adverse Events in Health Care
| No. | Domain | Patient Safety Topic |
| 1 | Structure | Organizational determinants and latent failures |
| 2 | Structure | Use of accreditation and regulation to advance patient safety |
| 3 | Structure | Safety culture |
| 4 | Structure | Inadequate training and education, manpower issues |
| 5 | Structure | Stress and fatigue |
| 6 | Structure | Production pressures |
| 7 | Structure | Lack of appropriate knowledge, availability of knowledge, transfer of knowledge |
| 8 | Structure | Having measures of patient safety |
| 9 | Structure | Devices, procedures without human factors engineering |
| 10 | Process | Errors in care through misdiagnosis |
| 11 | Process | Errors in care through poor test follow-up |
| 12 | Process | Errors in care: counterfeit/substandard drugs |
| 13 | Process | Errors in care: unsafe injection practices |
| 14 | Process | Bringing patients voices into patient safety |
| 15 | Outcomes | Adverse events and injuries due to medical devices |
| 16 | Outcomes | Adverse events due to medications |
| 17 | Outcomes | Adverse events due to surgical errors |
| 18 | Outcomes | Adverse events due to healthcare-associated infections |
| 19 | Outcomes | Adverse events due to unsafe blood products |
| 20 | Outcomes | Patient safety among pregnant women and newborns |
| 21 | Outcomes | Patient safety concerns among older adults |
| 22 | Outcomes | Adverse events due to falls in the hospital |
| 23 | Outcomes | Injury due to pressure sores and decubitus ulcers |
| SOURCE: Jha (2008). | ||
into three groups: structural factors, processes of care, and outcomes. Lead experts in each topic area described the basic epidemiology of the topic, how the issue impacts patient care, and knowledge gaps to be addressed through future research.
Findings from the work are striking and identify large gaps in current data to inform priority setting. The overarching message of the evidence is that unsafe medical care continues to cause substantial morbidity, mortality, and years of life lost—particularly in the developing world. The majority of work has examined hospital care in developed nations and found adverse events rates of approximately 10% (Brennan et al., 2004; Davis et al., 2002, 2003; Thomas et al., 2000; Vincent et al., 2001). While few data exist on the care delivered in developing and transitional nations, these epidemiological studies suggest similar rates of adverse events but higher morbidity and mortality compared to developed nations (Jha, 2008). Thus, the consequences of unsafe care in the developing world appear to be much greater. Many of these events are not only preventable, but also expensive. Yet, safety remains low on the policy agenda.
While there is strong evidence on poor clinical outcomes as a result of unsafe care in developed nations and a small but growing number of smaller studies in developing and transitional nations, knowledge on structural factors and processes in care is not nearly as robust. The findings of the report underscore the need to fill the large gaps in data to inform the design of solutions and track strategies for improvement. Notably, understanding how to best address safety in different settings, determining which solutions are exportable among nations, and assessing the cost-effectiveness of specific solutions will be critical to guide policy makers as they make important, difficult decisions on how to allocate limited resources to improve health across the globe. Without more data, formulating effective solutions will pose a substantial challenge.
The Global Burden of Disease
Building on the work of the report, the World Alliance for Patient Safety focused on quantifying the global burden of unsafe care. The global burden of disease is the metric used by WHO, policy makers, and funders to allocate global health resources. The fundamental ability to accurately calculate the global burden of diseases is dependent on the types of data available. These results have vast implications for how big of a priority patient safety is deemed.
To calculate the global burden of disease, the 10 major types of preventable events that were identified in the report on global patient safety were used (Table 8-2). Using existing data, the group then developed two new analytical models: (1) health burden, measured by disability-adjusted
TABLE 8-2 Adverse Event Conditions Used by WHO to Calculate the Global Burden of Disease
| Condition |
| Adverse drug events Venous thromboembolism complications Decubitus ulcers Falls in the healthcare setting Unsafe maternal/pregnancy care Hospital-acquired infections Surgical complications Adverse medical device events Unsafe blood products Unsafe injection practices Counterfeit medications |
| SOURCE: Jha (2008). |
life years (DALYs) lost (due to injury and mortality) and (2) economic burden, measured by the financial impact (i.e., increased length of stay, repeated surgeries) on healthcare systems and society. The models included the number of people at risk, rate of hospitalization, average age at the time of acquiring the condition, four clinical outcomes (death, short-term disability followed by long-term disability, short-term disability then full recovery, no or minimal disability), average duration of the condition, average direct costs related to care of condition per episode, and disability weights.
The findings were again powerful and indicate that unsafe care is one of the major causes of disability and death in the world. Initial estimates suggest that over 34 million adverse events in hospitals occur among the conditions examined (over 60 percent from developing and transitional countries), and that the global burden of unsafe care from these conditions may account for as many as 20 million DALYs lost per year (approximately 60 percent of which are from developing and transitional countries). The number of estimated DALYs lost due do unsafe care falls directly behind top major global causes of disability and death, such as lower respiratory infection (94.5 million DALYs), unipolar depression disorders (65.5 million DALYs), ischemic heart disease (62.6 million DALYS), and cerebrovascular disease (46.6 millions DALYs) (WHO, 2008a). However, unlike these conditions, much unsafe care is preventable. Furthermore, these results are likely to be conservative since not all types of adverse events were included in the calculations. Thus, designing and implementing successful interventions to curb unsafe care may be an important area to prioritize global health efforts.
While the models were based on the most current and comprehensive data available, the research methodology further highlighted the reality that there is a paucity of systematic data sources globally. Particularly in developing and transitional nations, there is extensive variability in the data. For example, rates of hospitalization among these nations ranged from 8 percent to 98 percent. While hospitalization estimates in developed nations were between 113 and 147 million, the estimates were between 111 and 469 million in developing and transitional countries. The data, still qualified in developing nations, only exists on the prevalence of injury (how often patients are injured in the hospital). The global burden of disease models requires more key data on patient demographics, the severity of disability, and injury duration. Until we have these data elements and more robust information infrastructure that facilitates the collection and analysis of these data, precise estimations to inform policy makers will be a major challenge.
WHO Resource-Poor Setting Initiative
Given the acute need for better data to help policy makers make decisions in poor, resource-lacking countries, WHO has begun thinking about identifying the minimum dataset needed in the developing world. Implementing comprehensive electronic health records and health information exchange infrastructure in the developing world is not a realistic strategy at the present date. Thus, WHO has convened an expert consensus group to identify the major causative structural factors (i.e., lack of protocols or systematic monitoring) that drive a few key patient safety issues and then determine a systematic method to collect the data elements hospitals need to overcome structural failings. This is an important initial step to obtaining the basic information that will help paint a broader picture on the scope of patient safety issues and understand how these issues may be resolved.
Conclusion
In summary, we find that the much of the developing and transitional world faces challenges similar to those of the United States and other high-income countries: ensuring the delivery of high-quality, safe care in an efficient way. While the issues of access to health care feel paramount to developing nations, ensuring access to safe, effective care is critically important. Our preliminary work suggests that millions of the world’s citizens—a majority in developing countries—are injured or killed due to unsafe health care. Information systems, whether they be rudimentary or advanced, are central to helping resource-poor nations develop an approach to improving patient safety, and building the trust of patients in the healthcare system in order to ensure that all of the world’s citizens have access to safe, effective care.
INFORMATICS AND THE FUTURE OF
INFECTIOUS DISEASE SURVEILLANCE
David L. Buckeridge, M.D., Ph.D., and John S. Brownstein, Ph.D.
McGill University
Advances in information technology are enabling dramatic changes in domestic and global infectious disease surveillance. Understanding the nature of these changes is critical to ensuring that existing and novel surveillance systems contribute effectively to disease control. In this paper, we describe how information technology is altering the surveillance landscape and identify how public health should harness these changes for effective disease control.
Traditional Domestic and International Surveillance Systems
Infectious disease surveillance has evolved over the last century to exploit many sources of information, but even where capacity is sufficient, systems based upon laboratory-confirmed diagnoses remain the preferred approach (Van Beneden and Lyndfield, 2010). Recent epidemics and pandemics, however, have highlighted the limited sensitivity and timeliness of laboratory-based systems. Since a case can be detected only if an infected person seeks medial care, sensitivity is limited by patterns of healthcare utilization. During the clinical encounter, sensitivity can be further reduced if a clinician does not order a laboratory test that can identify the organism under surveillance, or if a test is not routinely available.
The reporting of a laboratory-confirmed case of infection to a public health department is usually a manual process, which can take a week or longer to occur. Moreover, subsequent reporting between public health jurisdictions tends to follow a hierarchical pattern: a local health department informing a regional public health authority which then informs the national public health authority, a process that often takes 2 to 3 weeks (Birkhead et al., 1991; Jajosky and Groseclose, 2004; Jansson et al., 2004; Yoo et al., 2009). Finally, the national public health authority may inform the World Health Organization in accordance with the International Health Regulations (WHO, 2008b).
In the context where lab resources are constrained, systems for public health surveillance face similar limitations. Existing networks of traditional surveillance efforts—managed by health ministries, public health institutes, multinational agencies, and laboratory and institutional networks—have wide gaps in geographic coverage, capacity, and training, often resulting in poor and sometimes suppressed information flow.
Using Information Technology to Enhance Existing Systems
Advances in information technology are beginning to alter the landscape of infectious disease surveillance by addressing the limitations of traditional surveillance approaches. For example, large-scale telephone consultation lines that rely upon computerized decision algorithms (such as National Health Service Direct in the UK) attempt to direct patients to the appropriate level of clinical care (Snooks et al., 2009). Such streamlining of care may benefit laboratory-based surveillance by increasing the likelihood that those with diseases under surveillance seek care. Another application of information technology that may enhance existing surveillance systems is the use of decision support to prompt clinicians to order tests for conditions under surveillance (Lurio et al., 2010).
One of the more concerted attempts to apply information technology to modernize existing surveillance systems has aimed to automate the reporting of positive results from laboratories to public health departments. Evidence suggests that such automation can enhance sensitivity and improve the timeliness of reporting, reducing delays in initial reports from laboratories by 4 to 7 days (Effler et al., 1999; Overhage et al., 2008; Panackal et al., 2002; Ward et al., 2005). In the United States, considerable resources are being directed toward the acquisition of clinical information systems that support such electronic laboratory reporting (Blumenthal and Tavenner, 2010).
These applications of information technology have the potential to improve existing surveillance systems but they cannot resolve some of the most important limitations of surveillance. In resource-poor settings, they cannot address the issue of laboratory testing capacity. Even where laboratory resources are sufficient, improving test ordering and reporting does little to address the delays inherent in hierarchical reporting among public health jurisdictions after initial reports are received from laboratories.
Using Information Technology to Disrupt the Traditional Approach
In addition to enhancing existing surveillance systems, advances in information technology are also disrupting the traditional public health surveillance model by enabling new approaches to data sharing. Data are increasingly available from sources other than laboratories and these novel types of surveillance data are often shared outside of traditional public health channels. In contrast to the hierarchy that typifies reporting of laboratory-confirmed cases, data are increasingly shared more broadly, with decreased control over data sharing by governmental agencies.
The DiSTRIBuTE project2 is one example of an innovative approach to sharing surveillance data extracted from sources other than laboratories (Buckeridge et al., 2011). This project builds on the growing adoption of syndromic surveillance systems (Buehler et al., 2009), which allow public health departments to follow the reasons for visits to emergency departments (EDs) in their jurisdictions (Mandl et al., 2004). Although these ED data lack the specificity of laboratory-confirmed reports, they are sensitive, available immediately, and have been shown to correlate well with laboratory-confirmed reports for diseases such as influenza (Marsden-Haug et al., 2007). The DiSTRIBuTE project allows health departments with syndromic surveillance systems to rapidly share information from their systems. Over one-third of ED visits in the United States are now captured by the DiSTRIBuTE system, and information extracted from these data to support influenza surveillance are made publicly available with a delay of less that 72 hours for the majority of participating health departments (Buckeridge et al., 2011).
HealthMap is another example of using information technology to expand the scope of surveillance sources and free the flow of surveillance information. HealthMap harnesses and organizes the enormous amount of valuable epidemic intelligence found in web-accessible sources such as discussion sites, disease reporting networks, and news outlets (Freifeld et al., 2008). These resources provide current, highly local information about outbreaks—even from areas relatively invisible to traditional global public health efforts. These web-based data sources not only facilitate early outbreak detection, but also support increasing public awareness of disease outbreaks prior to their formal recognition (Brownstein et al., 2010).
A Renewed Science of Surveillance on the
Road to Effective Disease Control
Applications of information technology are enhancing existing systems and disrupting current surveillance models to make more information about infectious diseases available with less delay. Although some applications of information technology that influence infectious disease surveillance are under the control of the public health system, many are not. This reality is both exciting and challenging for the future of public health surveillance. It points to a future where disease information is available broadly and quickly, but raises the questions of how, and by whom, this information will be used to further effective disease control.
Public health workers use surveillance data to assess population health status and project the likely evolution of that status in the face of available
___________________
2 See http://www.ISDSDistribute.org (accessed January 14, 2011).
interventions (Buehler et al., 2009). To accomplish these tasks, data from different surveillance sources must be combined (Khan et al., 2010). Such combination could make the most of highly specific laboratory data, when available, and more sensitive and timely data from other sources. Combining data to support decision making, however, requires an understanding of the nature and quality of the data, something that is not always available for novel data sources.
While concern about the nature and quality of data is appropriate, public health authorities cannot and should not avoid novel sources of data and rest complacent with traditional models of surveillance. Instead, public health surveillance as a discipline must extend its theoretical and practical foundations to embrace the opportunities presented by information technology. In other words, a renewed science of disease surveillance is needed; one that starts from public health principles and embraces information technology enhancements as well as disruptive changes on the road to improved disease control (Thacker et al., 1989).
REFERENCES
Birkhead, G., T. Chorba, S. Root, D. Klaucke, and N. Gibbs. 1991. Timeliness of national reporting of communicable diseases: The experience of the National Electronic Telecommunications System for Surveillance. American Journal of Public Health 81:1313-1315.
Blumenthal, D., and M. Tavenner. 2010. The “meaningful use” regulation for electronic health records. New England Journal of Medicine 363(6):501-504.
Bonander, J., and S. Gates. 2010. Public health in an era of personal health records: Opportunities for innovation and new partnerships. Journal of Medical Internet Research 12(3):e33.
Brennan, T. A., L. L. Leape, N. M. Laird, L. Hebert, A. R. Localio, A. G. Lawthers, J. P. Newhouse, P. C. Weiler, and H. H. Hiatt. 2004. Incidence of adverse events and negligence in hospitalized patients: Results of the Harvard Medical Practice Study. Quality and Safety in Health Care 13(2):145-151; discussion 151-152.
Brownstein, J. S., C. C. Freifeld, E. H. Chan, M. Keller, A. L. Sonricker, S. R. Mekaru, and D. L. Buckeridge. 2010. Information technology and global surveillance of cases of 2009 H1N1 influenza. New England Journal of Medicine 362(18):1731-1735.
Buckeridge, D. L., J. S. Brownstein, W. B. Lober, D. R. Olson, M. Paladini, D. Ross, L. Finelli, T. Kass-Hout, and J. W. Buehler. 2011. The DiSTRIBuTE Project: Rapid sharing of emergency-department surveillance data during the 2009 influenza A/H1N1 pandemic. (Submitted).
Buehler, J. W., E. A. Whitney, D. Smith, M. J. Prietula, S. H. Stanton, and A. P. Isakov. 2009. Situational uses of syndromic surveillance. Biosecurity and Bioterrorism 7(2):165-177.
Davis, P., R. Lay-Yee, R. Briant, W. Ali, A. Scott, and S. Schug. 2002. Adverse events in New Zealand public hospitals: Occurrence and impact. New Zealand Medical Journal 115(1167):U271.
———. 2003. Adverse events in New Zealand public hospitals: Preventability and clinical context. New Zealand Medical Journal 116(1183):U624.
Delaney, B. C. 2008. Potential for improving patient safety by computerized decision support systems. Family Practice 25(3):137-138.
Dodd, B. 1997. An independent “health information bank” could solve data security issues. British Journal of Healthcare Computing and Information Management 14(8):33-35.
Effler, P., M. Ching-Lee, A. Bogard, M. Ieong, T. Nekomoto, and D. Jernigan. 1999. Statewide system of electronic notifiable disease reporting from clinical laboratories: Comparing automated reporting with conventional. Journal of the American Medical Association 282:1845-1850.
European Parliament and Council of the European Union. 2010. Directive 95/46/ec. http://eur-lex.europa.eu/LexUriServ/LexUriServ.do?uri=CELEX:31995L0046:en:NOT (accessed September 10, 2010).
Europe’s Information Society. 2009. I2010—A European information society for growth and employment. http://ec.europa.eu/information_society/eeurope/i2010/index_en.htm (accessed November 16, 2010).
Foster, I., and C. Kesselman. 2004. The Grid 2: Blueprint for a new computing infrastructure. Oxford, UK: Elsevier Science.
Freifeld, C. C., K. D. Mandl, B. Y. Reis, and J. S. Brownstein. 2008. HealthMap: Global infectious disease monitoring through automated classification and visualization of Internet media reports. Journal of the American Medical Informatics Association 15(2):150-157.
Fridsma, D. B., J. Evans, S. Hastak, and C. N. Mead. 2008. The BRIDG project: A technical report. Journal of the American Medical Informatics Association 15(2):130-137.
Garg, A. X., N. K. Adhikari, H. McDonald, M. P. Rosas-Arellano, P. J. Devereaux, J. Beyene, J. Sam, and R. B. Haynes. 2005. Effects of computerized clinical decision support systems on practitioner performance and patient outcomes: A systematic review. Journal of the American Medical Association 293(10):1223-1238.
Iakovidis, I., and O. Purcarea. 2008. eHealth in Europe: From vision to reality. Studies in Health Technology and Informatics 134:163-168.
Jajosky, R., and S. Groseclose. 2004. Evaluation of reporting timeliness of public health surveillance systems for infectious diseases. BMC Public Health 4(1):29.
Jansson, A., M. Arneborn, K. Skorlund, and K. Ekdahl. 2004. Timeliness of case reporting in the Swedish statutory surveillance of communicable diseases 1998-2002. In Scandinavian Journal of Infectious Diseases 36(11-12):865-872.
Jha, A. K. editor. 2008. Summary of the evidence on patient safety: Implications for research. Geneva, Switzerland: World Health Organization.
Kalra, D., T. Beale, and S. Heard. 2005. The openEHR Foundation. Studies in Health Technology and Informatics 115:153-173.
Khan, A. S., A. Fleischauer, J. Casani, and S. L. Groseclose. 2010. The next public health revolution: Public health information fusion and social networks. American Journal of Public Health 100(7):1237-1242.
Kostopoulou, O., J. Oudhoff, R. Nath, B. C. Delaney, C. W. Munro, C. Harries, and R. Holder. 2008. Predictors of diagnostic accuracy and safe management in difficult diagnostic problems in family medicine. Medical Decision Making 28(5):668-680.
Lurio, J., F. P. Morrison, M. Pichardo, R. Berg, M. D. Buck, W. Wu, K. Kitson, F. Mostashari, and N. Calman. 2010. Using electronic health record alerts to provide public health situational awareness to clinicians. Journal of the American Medical Informatics Association 17(2):217-219.
Mandl, K. D., J. M. Overhage, M. M. Wagner, W. B. Lober, P. Sebastiani, F. Mostashari, J. A. Pavlin, P. H. Gesteland, T. Treadwell, E. Koski, L. Hutwagner, D. L. Buckeridge, R. D. Aller, and S. Grannis. 2004. Implementing syndromic surveillance: A practical guide informed by the early experience. Journal of the American Medical Informatics Association 11(2):141-150.
Marsden-Haug, N., V. B. Foster, P. L. Gould, E. Elbert, H. Wang, and J. A. Pavlin. 2007. Code-based syndromic surveillance for influenzalike illness by International Classification of Diseases, ninth revision. Emerging Infectious Diseases 13(2):207-216.
Martin-Sanchez, F., I. Iakovidis, S. Nørager, V. Maojo, P. de Groen, J. Van der Lei, T. Jones, K. Abraham-Fuchs, R. Apweiler, A. Babic, R. Baud, V. Breton, P. Cinquin, P. Doupi, M. Dugas, R. Eils, R. Engelbrecht, P. Ghazal, P. Jehenson, C. Kulikowski, K. Lampe, G. De Moor, S. Orphanoudakis, N. Rossing, B. Sarachan, A. Sousa, G. Spekowius, G. Thireos, G. Zahlmann, J. Zvárová, I. Hermosilla, and F. J. Vicente. 2004. Synergy between medical informatics and bioinformatics: Facilitating genomic medicine for future health care. Journal of Biomedical Informatics 37(1):30-42.
OASIS (Organization for the Advancement of Structured Information Standards). 2005. eXtensible Access Control Markup Language (XACML) version 2.0. http://docs.oasisopen.org/xacml/2.0/access_control-xacml-2.0-core-spec-os.pdf (accessed September 10, 2010).
Ohmann, C., and W. Kuchinke. 2009. Future developments of medical informatics from the viewpoint of networked clinical research. Interoperability and integration. Methods of Information in Medicine 48(1):45-54.
Overhage, J. M., S. Grannis, and C. J. McDonald. 2008. A comparison of the completeness and timeliness of automated electronic laboratory reporting and spontaneous reporting of notifiable conditions. American Journal of Public Health 98(2):344-350.
Panackal, A. A., M. M’ikanatha N, F. C. Tsui, J. McMahon, M. M. Wagner, B. W. Dixon, J. Zubieta, M. Phelan, S. Mirza, J. Morgan, D. Jernigan, A. W. Pasculle, J. T. Rankin, Jr., R. A. Hajjeh, and L. H. Harrison. 2002. Automatic electronic laboratory-based reporting of notifiable infectious diseases at a large health system. Emerging Infectious Diseases 8(7):685-691.
Schade, C. P., F. M. Sullivan, S. de Lusignan, and J. Madeley. 2006. E-prescribing, efficiency, quality: Lessons from the computerization of UK family practice. Journal of the American Medical Informatics Association 13(5):470-475.
SHARE Collaboration. 2008. Share roadmap II. http://www.healthgrid.org/documents/pdf/SHARE_roadmap_long.pdf (accessed September 10, 2010).
Singh, H., E. J. Thomas, M. M. Khan, and L. A. Petersen. 2007. Identifying diagnostic errors in primary care using an electronic screening algorithm. Archives of Internal Medicine 167(3):302-308.
Snooks, H., J. Peconi, J. Munro, W. Y. Cheung, J. Rance, and A. Williams. 2009. An evaluation of the appropriateness of advice and healthcare contacts made following calls to NHS Direct Wales. BMC Health Services Research 9:178.
Speedie, S. M., A. Taweel, I. Sim, T. N. Arvanitis, B. Delaney, and K. A. Peterson. 2008. The Primary Care Research Object Model (PCROM): A computable information model for practice-based primary care research. Journal of the American Medical Informatics Association 15(5):661-670.
Thacker, S. B., R. L. Berkelman, and D. F. Stroup. 1989. The science of public health surveillance. Journal of Public Health Policy 10(2):187-203.
Thomas, E. J., D. M. Studdert, H. R. Burstin, E. J. Orav, T. Zeena, E. J. Williams, K. M. Howard, P. C. Weiler, and T. A. Brennan. 2000. Incidence and types of adverse events and negligent care in Utah and Colorado. Medical Care 38(3):261-271.
Van Beneden, C., and R. Lyndfield. 2010. Public health surveillance for infectious diseases. In Principles and practice of public health surveillance, edited by L. M. Lee, S. M. Teutsch, S. B. Thacker, and M. E. St. Louis. Oxford: Oxford University Press. Pp. 236-254.
Vincent, B., D. Kevin, and S. Tony. 2005. The healthgrid white paper. In From grid to HealthGrid: Proceedings of HealthGrid 2005. Oxford, UK: ISO Press. Pp. 249-321.
Vincent, C., G. Neale, and M. Woloshynowych. 2001. Adverse events in British hospitals: Preliminary retrospective record review. British Medical Journal 322(7285):517-519.
Ward, M., P. Brandsema, E. van Straten, and A. Bosman. 2005. Electronic reporting improves timeliness and completeness of infectious disease notification, the Netherlands, 2003. European Surveillance 10(1):27-30.
Warren, R., A. E. Solomonides, C. del Frate, I. Warsi, J. Ding, M. Odeh, R. McClatchey, C. Tromans, M. Brady, R. Highnam, M. Cordell, F. Estrella, M. Bazzocchi, and S. R. Amendolia. 2007. Mammogrid—a prototype distributed mammographic database for Europe. Clinical Radiology 62(11):1044-1051.
WHO (World Health Organization). 2008a. Global burden of disease: 2004 update. Geneva, Switzerland.
———. 2008b. International health regulations. http://www.searo.who.int/LinkFiles/International_Health_Regulations_IHR_2005_en.pdf (accessed January 14, 2011).
World Wide Web Consortium. 2004. SWRL: A Semantic Web Rule Language combining OWL and RULEML. http://www.w3.org/Submission/SWRL/ (accessed September 10, 2010).
———. 2009. OWL: Web Ontology Language—overview. http://www.w3.org/TR/owl-features/ (accessed September 10, 2010).
Yoo, H. S., O. Park, H. K. Park, E. G. Lee, E. K. Jeong, J. K. Lee, and S. I. Cho. 2009. Timeliness of national notifiable diseases surveillance system in Korea: A cross-sectional study. BMC Public Health 9:93.


























