1
Tempo, Mode, the Progenote, and the Universal Root
W. Ford Doolittle and James R. Brown
Simpson sought in Tempo and Mode in Evolution to explain large-scale variations in evolutionary rate and pattern apparent in the fossil record (Simpson, 1944). By tempo he meant "rate of evolution … practically defined as amount of morphological change relative to a standard," and by mode he meant "the way, manner, or pattern of evolution." For those of us concerned with the evolution of molecules rather than organisms, issues of tempo mostly have to do with the molecular clock, while questions about mode address mutational mechanisms and forces driving changes in gene and genome structure. In this article, we focus on the period of early cellular evolution, between the appearance of the first self-replicating informational macromolecule and the deposition of the first microfossils, by all accounts already modern cells (Schopf, 1994). We ask whether major shifts in predominant mode occurred during this period, and (since the answer is of course yes) whether we might actually come to know anything other than the vaguest generalities about these shifts.
W. Ford Doolittle is a Fellow of the Canadian Institute for Advanced Research and professor of biochemistry at Dalhousie University, Halifax, Nova Scotia. James R. Brown is a postdoctoral fellow in biochemistry at Dalhousie University, whose work is supported by the Medical Research Council of Canada.
Stages in the Evolution of the Cellular Information-Processing System
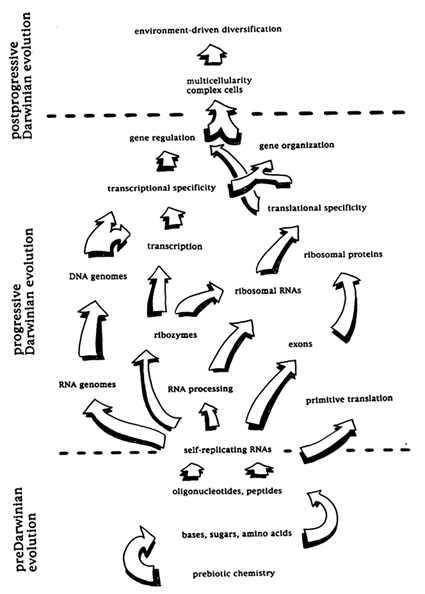
In Figure 1 we present a fanciful representation of the evolution of the information transfer system of modern cells and propose that it be seen as divisible into three phases, differing profoundly in both tempo and mode. The first (Figure 1 Bottom) would be accepted by all who speculate on the origin of Life as a period of preDarwinian evolution: without replication there are no entities to evolve through the agency of natural selection. We call the second period, between the appearance of the first self-replicating informational molecule and the appearance of the first "modern" cell, the period of progressive Darwinian evolution (Figure 1 Middle). "Progress" is of course an onerous concept in evolutionary theory (Ayala, 1988). Nevertheless, we submit that, as its uniquely defining feature or mode, this second phase witnessed the fixation of many mutations improving the accuracy, speed, and efficiency of information transfer overall and, thus, the adaptedness of cells (or simpler precellular units of selection) under almost any imaginable conditions. Nowadays (in the third period, that of postprogressive Darwinian evolution; Figure 1 Top), most mutations that are fixed by selection improve fitness only for specific environmental regimes. But earlier, when evolution did exhibit progress, selection forged successive generations of organisms (or simpler units) in which phenotype was more reliably coupled to genotype. Individuals from later in this period would have almost always outperformed their ancestors if placed in direct competition with them.
How could we hope to know anything about this ancient era of radically different tempo and mode? If divergences that established the major lineages of contemporary living things occurred before completion of the period of progressive Darwinian evolution, then we would expect that the information processing systems of these lineages would differ from each other—the earlier the divergence, the more profound the difference. That is, components of the replication, transcription, and translation machineries that were still experiencing progressive Darwinian evolution at the time of divergence should be differently refined or altogether separately fashioned (nonhomologous) in major lineages. Thus, comparisons between modern major groups (such as prokaryotes and eukaryotes) might lead to informed guesses about primitive ancestral states.
As an exemplary exercise, Benner and colleagues (Benner et al., 1993) inferred from the fact that archaebacteria, eubacteria, and eukaryotes produce ribonucleotide reductases that are not demonstrably homologous that their last common ancestor used a ribozyme for the reduction
of ribonucleotides. Benner's group also (and much more persuasively) concluded, from the evident homology of DNA (or RNA) polymerases in the three domains, that the transition from RNA to DNA genomes had itself already been made by that last common ancestor, whatever its residual reliance on ribozymology (Benner et al., 1989).
To make such inferences about past events through use of parsimony arguments to reconstruct common ancestors from knowledge of the different paths taken by descendants, we must know that the contemporary groups compared really did begin to diverge at an appropriately ancient date. In the rest of this review, we consider developments in our thinking about the relationships between basic kinds of living things primarily as they bear on this issue, asking if there is any reason to believe that the cenancestor was a progenote.
Of the two new terms introduced here, cenancestor is Walter Fitch's for "the most recent common ancestor to all the organisms that are alive today (Fitch and Upper, 1987)". Progenote is George Fox and Carl Woese's descriptor for "a theoretical construct, an entity that, by definition, has a rudimentary, imprecise linkage between its genotype and phenotype (Woese, 1987)"—a creature still experiencing progressive Darwinian evolution, in other words.
The Basic Kinds of Living Things
For more than a century, microbiologists suspected that bacteria, because of their small size and seemingly primitive structure, might differ fundamentally from animals, plants, and even fungi. The blue-green algae (now "cyanobacteria") might be intermediate, looking like bacteria but acting like plants. Chatton in 1937 (Chatton, 1937) and Stanier and van Niel in 1941 (Stanier and van Niel, 1941) proposed that these two groups share a common cellular organization distinguishing them as prokaryotes from the rest of the living world, or eukaryotes. A clear statement of the differences, however, required further work in biochemistry, genetics, and cellular ultrastructure. By 1962, Stanier and van Niel (Stanier and van Niel, 1962) were prepared to define prokaryotes in terms of the specific features they shared as well as the eukaryotic characteristics they lacked. They wrote that:
… the principal distinguishing features of the prokaryotic cell are:
- absence of internal membranes which separate the resting nucleus from the cytoplasm, and isolate the enzymatic machinery of photosynthesis and respiration in specific organelles;
- nuclear division by fission, not by mitosis, a character possibly related to the presence of a single structure which carries all the genetic information of the cell; and
- the presence of a cell wall which contains a specific mucopeptide as its strengthening element.
By 1970, Stanier could confidently state that
… advancing knowledge in the domain of cell biology has done nothing to diminish the magnitude of the differences between eukaryotic and prokaryotic cells that could be described some ten years ago: if anything, the differences now seem greater (Stanier, 1970).
But, cautiously endorsing Lynn Margulis' assertion that eukaryotic cells are themselves the result of the fusion of separate (prokaryotic) evolutionary lineages (Margulis, 1970), he went on to note that
… the only major links [between the two cell types] which have emerged from recent work are the many significant parallelisms between the entire prokaryotic cell and two component parts of the eukaryotic cell, its mitochondria and chloroplasts.
This linkage has since been amply supported by molecular sequence data (Gray and Doolittle, 1982), and the endosymbiont hypothesis for the origin of eukaryotic organelles of photosynthesis and respiration has become a basic tenet of the contemporary evolutionary consensus.
Together, the prokaryote/eukaryote dichotomy and the endosymbiont hypothesis for the origin of mitochondria and chloroplasts informed and (no doubt) constrained the biology and molecular biology of the 1960s, 1970s, and early 1980s, providing the framework within which all of the results of biochemists, geneticists, and evolutionists were interpreted (Figure 2). In typical text books from this era, genes in Escherichia coli are compared and contrasted to their counterparts in yeast, mouse, and man, with differences interpreted either in terms of the relatively advanced and complex state of the latter or the admirably streamlined features of the former. The paradigm has been extraordinarily fruitful: without such a grand scheme for organizing our knowledge of cell and molecular biology, we would have become lost in the details. It also seems safe to say that, for the organisms studied by most molecular biologists in those decades, this view of things is substantially correct and invaluable in interpreting the differences in the information-transfer systems of prokaryotes and eukaryotic nuclei, chloroplasts, and mitochondria.
As well, this view was easily consistent with the most straightforward interpretation of the fossil record. As reviewed by Schopf and Knoll elsewhere in this volume, unquestionable prokaryotes, by all available measures indistinguishable from modern cyanobacteria, appeared more than 3.5 billion years ago (Schopf, 1994; Knoll, 1994). Fossils that are undeniably eukaryotic are not seen for another 1 to 1.5 billion years, ample time for the symbioses required by Margulis.
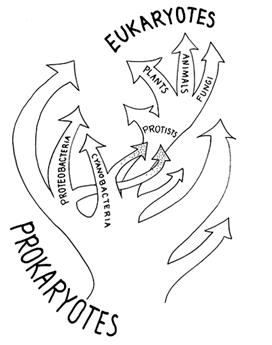
Figure 2 Prevalent evolutionary view between 1970 and 1977. The eukaryotic nuclear lineage arose from within the already characterized prokaryotes (eubacteria, perhaps a mycoplasma). Mitochondria descend from endosymbiotic proteobacteria, and plastids descend from endosymbiotic cyanobacteria.
The Woesian Revolution
The consensus represented by Figure 2 rested on comparative ultrastructural, biochemical, and physiological data and on a modest accumulation of primary (protein) sequence information, mostly from cytochromes and ferredoxins. In 1978, Schwartz and Dayhoff summarized this information and the then even-more-limited data from ribosomal RNA (rRNA)—in particular, 5S rRNA (Schwartz and Dayhoff, 1978). The endosymbiotic nature of organelles was well supported, but the origin of the nuclear genome (that is, the genome of the host for these endosymbioses) remained a mystery. A grand reconstruction of all of the main events of evolution with a single molecular chronometer was called for.
Such a grand reconstruction was the goal of Woese, who had begun, in the late 1960s, to assemble catalogs of the sequences of the oligonucleotides released by digestion of in vivo-labeled 16S rRNA with T1
ribonuclease. Comparing catalogs from different bacteria (scoring for presence or absence of identical oligonucleotides) by methods of numerical taxonomy allowed the construction of dendrograms showing relationships between them (Woese and Fox, 1977). Methods have been updated, cataloging giving way to reverse-transcriptase sequencing of rRNA, and this in turn to cloning (and now PCR cloning) of DNAs encoding rRNA (rDNAs). The data bases presently contain partial or complete sequences for some 1500 small-subunit rRNAs from prokaryotes and a rapidly growing collection of eukaryotic cytoplasmic small-subunit sequences, which track the evolutionary history of the nucleus (Olsen et al., 1994).
The rRNA data support the consensual picture represented in Figure 2 in many important ways. Such data not only confirm that chloroplasts and mitochondria descend from free-living prokaryotes but also show that the former belong close to (perhaps within) the cyanobacteria, while the latter derive from the alpha subdivision of the purple bacteria (proteobacteria). These data also establish relationships within the bacteria that are sensible in terms of advancing knowledge of prokaryotic biochemical and ecological diversity and often congruent with more traditional classification schemes, at least at lower taxonomic rank. However, there were two major surprises, both announced by Woese and colleagues in 1977 (Woese and Fox, 1977; Fox et al., 1977).
The first was that the eukaryotic nuclear lineage, as tracked by (18S) cytoplasmic small-subunit rRNA, was not demonstrably related to any specific, previously characterized prokaryotic lineage (Figure 3). This was not expected: the endosymbiont hypothesis saw the endosymbiotic host arising within the bacteria, the descendant of some otherwise typical prokaryote that had lost its cell wall and acquired the ability to engulf other cells. Differences in primary sequence between eukaryotic and prokaryotic small-subunit rRNAs also bespoke differences in secondary structure, consonant with the known differences in size (80S versus 70S), ribosomal protein content (75–90 polypeptides rather than 50–60), and function (initiation through ''scanning" rather than base-pairing via the Shine-Dalgarno sequence, unformylated rather than formylated initiator tRNA).
Because of these differences, Woese argued that the ribosome of the last common ancestor of bacteria and eukaryotes (their nuclear-cytoplasmic part, that is) was itself a primitive ribosome, a structure still experiencing progressive Darwinian evolution. He ventured (Woese, 1982) that the same might be said for other components of this cenancestral information processing system and that:
… in such a progenote, molecular functions would not be of the complex, refined nature we associate with functions today. Thus subsequent evolution would alter functions mainly in the sense of refining them. In this way, the
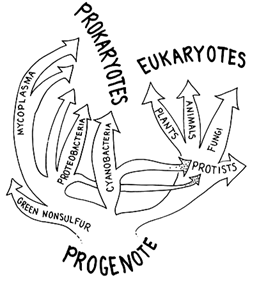
Figure 3 Implications of the rRNA data of Woese and Fox (1977). Prokaryotic (eubacterial) lineages from which the eukaryotic nuclear lineage was thought to have evolved were entirely separate from that lineage. Distinct properties of rRNAs suggested that the ribosome of the last common ancestor was a primitive ribosome and that the last common ancestral cell was a primitive "progenote," still experiencing progressive Darwinian evolution.
molecular differences among the three major groups would be in refinements of functions that occurred separately in the primary lines of descent, after they diverged from the universal ancestor.
In other words, the cenancestor was a progenote—one of the series of ancestral forms in which the phenotype-genotype coupling was actively evolving, and we might learn about progressive Darwinian evolution by comparing prokaryotic and eukaryotic (nuclear) molecular biology. Woese went on:
… it is hard to avoid concluding that the universal ancestor was a very different entity from its descendants. If it were a more rudimentary sort of organism, then the tempo of its evolution would have been higher and the mode of its evolution highly varied, greatly expanded (Woese, 1987).
This view came to play a dominant role in the molecular biology and evolutionary microbiology of the 1980s and early 1990s. The prokaryote/eukaryote dichotomy remained, but as a vertical split, separating living things into two camps from the very beginning rather than marking a more recent but crucial transition in the grade of cellular
organization. The inference that the cenancestor was a rudimentary being gave aid and comfort to those of us who had always doubted that the profound differences in gene and genome structure between eukaryotic nuclei and prokaryotes were improvements or advancements wrought in the former after their emergence from among the latter. Eukaryotic nuclear genomes are after all very messy structures, with vast amounts of seemingly unnecessary "junk" DNA, difficult-to-rationalize complexities in mechanisms of transcription and mRNA modification and processing, and needless scattering of genes that often in prokaryotes would be neatly arranged into operons. It might be easiest to see nuclear genomes as in a primitive state of organization, which prokaryotes, by dint of vigorous selection for economy and efficiency ("streamlining"), have managed to outgrow.
Such a view gained credence from and lent credence to the still popular although increasingly untenable "introns early" hypothesis or "exon theory of genes" (Doolittle, 1991). In brief, the notion here is that (i) the first self-replicators were small RNAs, which became translatable into small peptides; (ii) such "minigenes" came together to form the (RNA) ancestors of modern genes, introns marking the sutures; and (iii) the subsequent history of introns has been one of loss: streamlining has removed them entirely from the genes of prokaryotes but has been less effective in eukaryotes for a variety of reasons (less intense selection, lack of transcription-translation coupling as a driving force).
The second surprise from the rRNA data is depicted in Figure 4. In addition to showing the profound division between eukaryotes (their nuclei) and prokaryotes just discussed, these data identified two deeply diverging groups, two "primary kingdoms" within the prokaryotes. Woese and Fox called the first, which included E. coli and other proteobacteria, Bacillus subtilis, mycoplasma, the cyanobacteria, and indeed all prokaryotes about which we had accumulated any extensive biochemical or molecular genetic information, the "eubacteria" (Woese and Fox, 1977). It was these organisms that Stanier and van Niel had in mind when defining the prokaryote-eukaryote dichotomy in the 1950s and 1960s and on which most of us still fashion our beliefs about prokaryotes. The second primary kingdom, the "archaebacteria ," included organisms that, although certainly not unknown to microbiologists, had been little studied at the cellular and molecular level, and whose inclusion within the prokaryotes therefore rested at that time on only the most basic of criteria (absence of a nucleus).
Archaebacteria are organisms of diverse morphology and radically different phenotypes, including the obligately anaerobic mesophilic methanogens, the aerobic and highly salt-dependent extreme halophiles, the amazing (because capable of growth up to at least 110°C)

Figure 4 Further implications of the rRNA data (Woese and Fox, 1977). A third group, the archaebacteria, seemed as distant evolutionarily from eubacteria and eukaryotes as these were from each other.
extreme thermophiles, and still completely uncharacterized and unseen meso- or psychrophiles, which are related to the extreme thermophiles and known only from PCR products amplified from the open ocean (DeLong, 1992). Uniting them are a number of basic characters unrelated to rRNA sequence and more than adequate to support their taxonomic and phylogenetic unity in spite of this diversity. These include unique isopranyl ether lipids (and the absence of acyl ester lipids found in eubacteria and eukaryotes); characteristic genetic organization, sequence, and function of RNA polymerase subunits; structural and functional characteristics of ribosomes and modification patterns of tRNAs; varied but unique cell-envelope polymers; and distinctive antibiotic sensitivities and insensitivities (Zillig et al., 1993).
Rooting the Universal Tree
Woese felt that the differences between archaebacteria and either eubacteria or eukaryotes were of a sufficiently fundamental nature to indicate that all three primary kingdoms must have begun to diverge during the period of progressive evolution from a progenote. But there was no way to decide the order of branching—whether the first divergence in the universal tree separated (i) eubacteria from a line that was to produce archaebacteria and eukaryotes, or (ii) a proto-eukaryotic
lineage from a fully prokaryotic (eubacterial and archaebacterial) clade, or (iii) the (the third and least popular possibility) archaebacteria from eukaryotes and eubacteria.
There is in fact in principle no way to decide this or to root such a universal tree based only on a collection of homologous sequences. We can root any sequence-based tree relating a restricted group of organisms (all animals, say) by determining which point on it is closest to an "outgroup" (plants, for example). But there can be no such organismal outgroup for a tree relating all organisms, and the designation of an outgroup for any less-embracing tree involves an assumption, justifiable only by other unrelated data or argument. Alternatively, we might root a universal tree by assuming something about the direction of evolution itself: Figure 2 for instance is rooted in the belief that prokaryotic cellular organization preceded eukaryotic cellular organization. But in fact the progenote hypothesis itself is such an assumption about the direction of evolution: we cannot use it to prove its own truth. We must establish which of the three domains diverged first by some other method—unrelated to either outgroup organisms or theories about primitive and advanced states—before we can start to use three-way comparative studies to make guesses about the common ancestor.
A solution to this problem was proposed and implemented by Iwabe and colleagues (Iwabe et al., 1989), in 1989. Although there can be no organism that is an outgroup for a tree relating all organisms, we can root an all-organism tree based on the sequences of outgroup genes produced by gene duplication prior to the time of the cenancestor. The reasoning is as follows. Imagine such an ancient gene duplication producing genes A and A', both retained in the genome of the cenancestor and all descendant lineages (Figure 5). Then either A or A' sequences can be used to construct unrooted all-organism trees, and the A tree can be rooted with any A' sequence, and the A' tree can be rooted with any A sequence. As well, there is a built-in internal check, because both trees should have the same topology!
What Iwabe et al. needed, then, were sequences of gene pairs that (because all organisms have two copies) must be the product of a precenancestral gene duplication and for which eubacterial, archaebacterial, and eukaryotic versions were known. Two data sets met their criteria—the α and β subunits of F1 ATPases and the translation elongation factors EF-1α (Tu) and EF-2 (G). With either data set, rooted trees showing archaebacteria and eukaryotic nuclear genomes to be sister groups were obtained; eubacteria represented the earliest divergence from the universal tree (Figure 6).
The archaebacteriological community was already primed to accept this conclusion. At the very first meeting of archaebacterial molecular
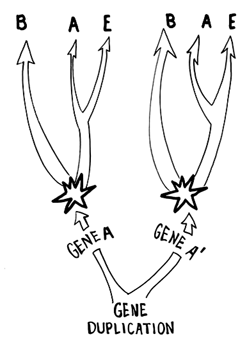
Figure 5 Rooting method of Iwabe and coworkers (1989). Two unrooted trees are constructed for eubacteria (B), archaebacteria (A), and eukaryotes (E), one with sequences of A genes and one with sequences of A' genes. A and A' genes are products of a gene duplication that must have predated the time of the last common ancestor because both are found in B, A, and E. The A tree can be rooted with A' sequences and vice versa, so that the cenacestor (the universal root) is in the position shown by the star for either tree.
biologists in Munich in 1981, one could sense a general feeling that archaebacteria were somehow "missing links" between eubacteria and eukaryotes. Zillig in particular stressed the (still-supported) eukaryote-like structural and functional characteristics of archaebacterial RNA polymerases (Zillig et al., 1982). In the subsequent 7 or 8 years, further gene sequences for proteins of the information-transfer system (ribosomal proteins, DNA polymerase) that looked strongly eukaryote-like had appeared. Although not rootable, these data too seemed to support a specific archaebacterial/eukaryotic affinity (Ramirez et al., 1993).
In 1990, Woese, Kandler and Wheelis incorporated the Iwabe rooting in a new and broader exegesis on the significance of the tripartite division of the living world (Woese et al., 1990). This treatment elevated the rank of the three primary kingdoms to "domains" (since kingdom
status was already well accepted for animals, plants, and fungi within the eukaryotes) and renamed them Bacteria, Archaea, and Eucarya. There were immediate and strong complaints from key figures in the evolutionary community, principally Lynn Margulis, Ernst Mayr and Tom Cavalier-Smith (Margulis and Guerrero, 1991; Mayr, 1990; Cavalier-Smith, 1992).
The objections touch many of the usual bases in evolutionary debates. Strict cladists would applaud the removal of "bacteria" from the name of the archaea, for instance, and would agree that the term "prokaryote" should not be used as a clade name because it describes a paraphyletic group. However, Woese and colleagues proposed the renaming not from cladist scruples but because of their belief in the profound nature of the phenotypic differences between archaebacteria and eubacteria. Mayr is no cladist either, but as a "gradist" he sees the change in cellular
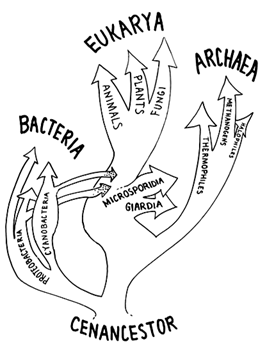
Figure 6 Currently accepted (Iwabe) rooting and renaming by Woese and collaborators of three primary kingdoms as domains (1990). Microsporidia and Giardia are archezoans thought to have diverged from the rest of the eukaryotic nuclear lineage before the acquisition of mitochondria or plastids through endosymbiosis.
grade represented by the prokaryote → eukaryote cellular transition as the major event in cell evolution. In lodging his objections to the paper of Woese, Kandler and Wheelis, he writes:
… as important as the molecular distance between the Archaebacteria and Eubacteria may seem to a specialist, as far as their general organization is concerned, the two kinds of prokaryotes are very much the same. By contrast, the series of evolutionary steps in cellular organization leading from the prokaryotes to the eukaryotes, including the acquisition of a nucleus, a set of chromosomes and the acquisition, presumably through symbiosis, of various cellular organelles (chloroplasts, mitochondria and so on) results in the eukaryotes in an entirely new level of organization … (Mayr, 1990).
Tom Cavalier-Smith (Cavalier-Smith, 1992) echoes this view:
Woese has repeatedly and mistakenly asserted that his recognition and firm establishment of the kingdom Archaebacteria (certainly a great and important breakthrough) invalidates the classical distinction between prokaryotes and eukaryotes. But as archaebacteria fall well within the scope of prokaryotes and bacteria as classically defined, it does nothing of the kind.
The questions that have to do with data rather than philosophy are: (i) what and how many traits distinguish the domains from each other (or betray a closer affinity between any two), (ii) how "fundamental" are these traits, and (iii) are such traits universally present within one (or two) groups and universally absent from the other(s) or is there in reality more of a mixing. For all the richness of our understanding of individual aspects of the biology of individual organisms, we are still very much in the dark, especially for answers to the second and third questions. Only recently, for instance (Cavalier-Smith, 1993), have we come to realize that archezoa (primitively amitochondrial eukaryotes) have 70S ribosomes, with rRNAs of the sizes and classes found in prokaryotes (archaebacteria or eubacteria). We know very little about possible forerunners of cytoskeletal proteins and functions in archaebacteria, although there have long been hints of such (Stein and Searcy, 1981). Even the eubacteria have not been plumbed in depth—newly discovered deeply branching lineages like Aquifex and the Thermotogales remain almost completely unknown in molecular or biochemical terms.
Implications of the Rooting for an Understanding of Tempo and Mode in Early Cellular Evolution
The Iwabe rooting and the renaming of the three domains as Bacteria, Archaea, and Eukarya (Figure 6) have found, in spite of these philosophical concerns, wide acceptance in the last 3 or 4 years. Together with increasing general understanding of gene and genome structure and function in the archaebacteria, the rooting has unavoidable impli-
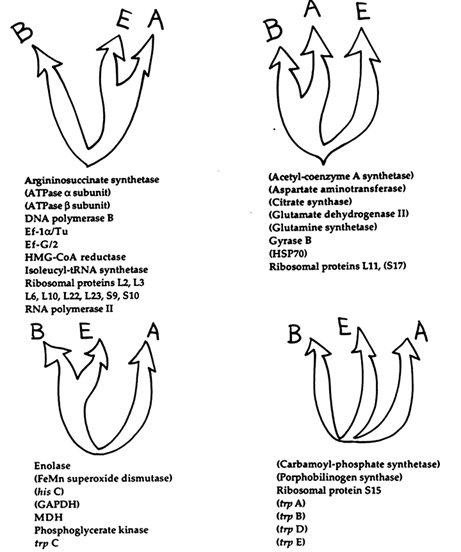
Figure 7 Categorization of proteins according to support for four possible rooted universal-tree topologies. Proteins were assigned to a particular topology based on the two domains determined to be closest after calculation of interdomain distances. Interdomain distances were estimated from the means of multiple pairwise comparisons between different species of each domain by using the program PROTDIST of the PHYLIP version 3.5 package (James R. Brown and W. Ford Doolittle, in preparation). For protein data sets shown in parentheses, at least one sequence did not support the monophyletic groups (B, A, and E) expected from rRNA data, according to the neighbor-joining method.
cations concerning the nature of the cenancestor and the possibility of learning about the period of progressive Darwinian evolution.
For instance, a specific lesson can be drawn from work in our laboratory (Cohen et al., 1992; Lam et al., 1990). The halophilic archaebacterium Haloferax volcanii was shown, by a variety of physical and genetic techniques, to have a genome made up of a large circular DNA of 2.92 million base pairs (Mbp) and several smaller but still sizeable molecules at 690, 442, 86, and 6 kbp. Of 60 or 70 genes known from cloned and sequenced fragments or through mutants, all but a doubtful 1 mapped to the 2.9 Mbp circle, which we thus called the chromosome, considering it similar to eubacterial chromosomes. There may of course be only so many ways to assemble a small genome: more telling is the fact that genes on this chromosome, and in thermophilic and methanogenic archaebacteria as well, are often organized into operons—cotranscribed and coordinately regulated clusters of overlapping genes controlling biochemically related functions. This too might be dismissed as a convergent or coincidental "eubacterial" feature (operons being unknown in eukaryotes), but the finding of tryptophan operons in Haloferax and in a methanogen and in the thermophile Sulfolobus (Meile et al., 1991; Tutino et al., 1993) seems more than coincidental, since clustering of tryptophan biosynthetic genes is almost universal among eubacteria. Most compelling of all are ribosomal protein gene clusters. In the L11-L10 clusters and the spectinomycin, S10, and streptomycin operons, 4 of 4, 11 of 11, 8 of 8, and 3 of 3 ribosomal protein genes are linked in the very same order (Ramirez et al., 1993) in E. coli and in the archaebacteria that have been looked at (often including a halophile, a methanogen, and a thermophile). These remarkable organizational similarities cannot be mere coincidence and are most unlikely to reflect convergence, since there is no clear reason why the genes must be linked in these precise orders. In fact, gene order is conserved even when positions of promoters (and hence units of coordinate regulation) are not. The last common archaebacterial/eubacterial ancestral genome must have had operons just like this and likely was very much like the present E. coli or Haloferax genomes in other specific and general respects (including origins and mechanism of replication, and so forth). If the Iwabe rooting is right, the last common archaebacterial/eubacterial ancestor is the last common ancestor of all Life. The genome of this cell, the cenancestor, would have been—as far as its organization is concerned—remarkably like that of a modern eubacterium, and we would have no hope of recreating the period of progressive Darwinian evolution by the comparative method.
There is a consolation, however, if this is true. We can then more surely say that the eukaryotic nuclear genome has become drastically
disorganized (or reorganized) since its divergence from its more immediate archaebacterial ancestor. As well, other characteristic features of eukaryotic nuclear molecular biology, such as multiple RNA polymerases and complex mRNA processing and intron splicing, must have appeared since this divergence.
We simply do not know how soon after the nuclear divergence these changes were wrought. The eukaryotes whose molecular biology we understand well—animals and fungi—are part of what has been called ''the crown" (Sogin, 1991) of the eukaryotic subtree (Figure 6). Very few genes have been cloned from protists diverging below the trypanosomes, and virtually nothing is known about their expression. It would not be foolish, if the Iwabe rooting holds, to anticipate that some diplomonads or microsporidia, which are thought to have diverged from the rest of the eukaryotes before the mitochondrial invasion, will turn out to have operons. Hopes of finding out just how archaebacteria-like such archezoal eukaryotic genomes are have now captured the interests and energies of several laboratories.
But Is the Rooting Right?
In a sense this new direction is an old one. Once again, we are examining the prokaryote -> eukaryote transition. Once again, as in Figure 2, we see the eukaryotic nuclear genome as the highly modified descendant of an already well-formed prokaryotic genome. The difference is that the immediate prokaryotic ancestors of the eukaryotic nuclear-cytoplasmic component are cells of a type we did not know when we first adopted the view shown in Figure 2. How we feel about the importance and novelty of this Hegelian outcome may depend on the side we take in the clade versus grade (Woese versus Mayr and Cavalier-Smith) debate discussed above. More to the point, however, is the possibility that we have accepted the Iwabe rooting, and consequently its implications for the modernity of the cenancestor and the radical remaking of the nuclear genome, too quickly and too uncritically. Iwabe and colleagues' data set included only one archaebacterial ATPase subunit pair (from Sulfolobus), only one elongation factor pair (Methanococcus), and a very limited representation of eubacterial sequences. As Gogarten (Hilario and Gogarten, 1993), and Forterre and his coworkers have recently and persuasively argued, both data sets can be questioned (Forterre et al., 1993). There is increasing evidence for multiple gene duplication events in the history of the ATPase genes, and it is difficult to distinguish orthologues (descendant from the same cenancestral α or β subunit gene) from paralogs (descendants of more distant homologs produced by gene duplication before the cenancestor). For the elonga-
tion factors, the alignment between EF1-α/Tu types and EF-2/G species, on the correctness of which the accuracy of the rooting absolutely depends, is highly problematic. More data for precenancestral gene duplications are sorely needed.
Along with the ATPase and elongation factor gene duplication analyses, it has become common to stress the similarity in sequence of archaebacterial and eukaryotic RNA polymerase subunits or (certain) ribosomal proteins. These indeed have shown a close archaebacterial/eukaryotic relationship by a variety of measures (Zillig et al., 1993). A broader survey of homologous genes for which readily alignable sequences are available for at least one species of each of the three domains is presented as Figure 7. (Eukaryotic nuclear genes suspected of being more recent acquisitions from bacterial endosymbiosis and extensively polyphyletic gene data sets are not shown.) In this figure, mean interdomain distances were used to construct midpoint rooted trees. The Iwabe tree is the most frequent among them, but not significantly so, and of course midpoint rootings can be correct only with constant molecular clocks.
So we must continue to remain open. If the currently accepted rooting were wrong, then an archaebacterial/eubacterial sisterhood seems the next most likely possibility, given the remarkable similarity in genetic organization between these two prokaryotic domains. The cenancestor could (again) be seen as a more primitive cell. Although it would have to possess all of those biochemical features known to be homologous in archaebacteria, eubacteria, and eukaryotes now (DNA genome, DNA polymerases, RNA polymerases, two-subunit ribosomes, the "universal code," most of metabolism, and many features of cell-cycle and growth regulation), we are free to see its genome as eubacteria-like, eukaryote-like, or something altogether different still (Woese, 1982; Woese, 1987). The fluid exchange of genes between lineages imagined by Woese in his early descriptions of the progenote remains possible.
Where Next?
The root of the universal tree is still "up in the air," and we don't know as much about the cenancestor as we had hoped. Why is this? One possibility is that we are pushing molecular phylogenetic methods to their limits: although we have reasonable ways of assessing how well any given tree is supported by the data on which it is based, methods for determining the likelihood that this is the "true tree" are poorly developed. Another is hidden paralogy—gene duplication events (of which there are only scattered detected survivors, different in different lineages) are fatal to the enterprise of phylogenetic reconstruction.
A mammalian tree drawn on the basis of myoglobin sequences from some species and hemoglobin sequences from others would be accurate as far as the molecules (which are all homologues) are concerned, but would be seriously wrong for the organisms. A third possibility, formally identical to paralogy in its baleful consequence for tree construction, is lateral (horizontal) gene transfer. Certainly such transfer has occurred within and between domains, early and late in their evolution (Smith et al., 1992). Zillig and Sogin (Zillig et al., 1993; Sogin, 1991) have drawn (quite different) scenarios in which extensive lateral transfer is invoked to explain the multiplicity of trees shown in Figure 7, each of which can then be taken at face value.
What renders all such attempts to resolve the current dilemma unnecessary and dangerously premature is the certainty that we will soon have enormously many more data. Total genome sequencing projects are under way for several eubacteria (E. coli, B. subtilis, a mycoplasma, and two mycobacteria), several archaebacteria (including Sulfolobus solfataricus ), and, of course, a number of "crown" eukaryotes seen as more direct models for the human genome. Instead of at most three dozen data sets with representative gene sequences from all three domains, we should have 3000. If the data in aggregate favor a single tree, this should be apparent. If there have been lateral transfers of related or physically linked genes, then we might be able to see them. If transfer has so scrambled genomes that we can no longer talk sensibly about the early evolution of cellular lineages but only of lineages of genes, then that too should be apparent, as would the need to change the very language with which we address an evolutionary process so radically different in both tempo and mode.
We should not allow our current confusion about the root to discourage us, and it is heartening to remember how far we have come. The prokaryote-eukaryote distinction has replaced that between animals and plants, and although we may no longer see that distinction as clearly as Stanier and van Niel thought they did, it is because we know more about the diversity of microbes; we will never go back to a world of just animals and plants. Similarly, the endosymbiont hypothesis for the origin of mitochondria and chloroplasts is as firmly established as any fact in biology; we will not return to the belief in direct filiation (bacteria -> cyanobacteria -> algae -> all other eukaryotes) which preceded it. As for the archaebacteria, although there remains some doubt as to their "holophyly" (thermophiles may be especially close to eukaryotes) and legitimate debate over the philosophical and biological implications of their existence for the meaning of the word "prokaryote," we will never again see these fascinating creatures scattered
taxonomically among the bacteria, as uncertain relatives of known eubacterial groups.
Methodologically, rRNA seems unlikely ever to lose pride of place as the most reliable molecular chronometer: Woese's original choice of this universally essential, functionally conservative and slowly evolving species was well justified. At the same time, protein data will increasingly supplement rRNA sequences—rRNAs may mislead us when they show base compositional biases, and there is anyway no single molecule which defines a cellular lineage, once lateral transfer is admitted. Molecular evolution is maturing, which means that the arguments of molecular evolutionists are becoming more pluralistic and subtler. We should welcome this, and the dialectic which assures that evolutionary theories are rarely wholely overthrown but instead are incorporated in unexpected ways and with unanticipated benefits into succeeding generations of biological thinking.
Summary
Early cellular evolution differed in both mode and tempo from the contemporary process. If modern lineages first began to diverge when the phenotype-genotype coupling was still poorly articulated, then we might be able to learn something about the evolution of that coupling through comparing the molecular biologies of living organisms. The issue is whether the last common ancestor of all life, the cenancestor , was a primitive entity, a progenote, with a more rudimentary genetic information-transfer system. Thinking on this issue is still unsettled. Much depends on the placement of the root of the universal tree and on whether or not lateral transfer renders such rooting meaningless.
Work in this laboratory described in this manuscript is supported by the Medical Research Council of Canada, of which agency J.R.B. is also a Postdoctoral Fellow. W.F.D. is a Fellow of the Canadian Institute for Advanced Research.
References
Ayala, F. J. (1988) Can "Progress" be defined as a biological concept? In Evolutionary Progress, ed. Nitecki, M. H. (Univ. Chicago Press, Chicago), pp. 75–96.
Benner, S. A., Cohen, M. A., Gonnet, G. H., Berkowitz, D. B. & Johnsson, K. P. (1993) Reading the palimpsest: contemporary biochemical data and the RNA world. In The RNA World, eds. Gesteland, R. F. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 27–70.
Benner, S. A., Ellington, A. D. & Tauer, A. (1989) Modern metabolism as a palimpsest of the RNA world. Proc. Natl. Acad. Sci. USA 86, 7054–7058.
Cavalier-Smith, T. (1992) Bacteria and eukaryotes. Nature (London) 356, 570.
Cavalier-Smith, T. (1993) Kingdom Protozoa and its 18 phyla. Microbiol. Rev. 57, 953–994.
Chatton, E. (1937) Titres et Travauz Scientifiques (Setes, Sottano, Italy).
Cohen, A., Lam, W. C., Charlebois, R. L., Doolittle, W. F. & Schalkwyk, L. C. (1992) Localizing genes on the map of the genome of Haloferx volcanii, one of the archaea. Proc. Natl. Acad. Sci. USA 89, 1602–1606.
DeLong, E. F. (1992) Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89, 5685–5689.
Doolittle, W. F. (1991) The origins of introns. Curr. Biol. 1, 145–146.
Fitch, W. M. & Upper, K. (1987) The phylogeny of tRNA sequences provides evidence for ambiguity reduction in the origin of the genetic code. Cold Spring Harbor Symp. Quant. Biol. 52, 759–767.
Forterre, P., Benanchenhou-Lahfa, N., Canfalonieri, F., Duguet, M., Elie, C. & Labedan, B. (1993) The nature of the last universal ancestor and the root of the tree of life: still open questions. BioSystems 28, 15–32.
Fox, G. E., Magrum, L. J., Batch, W. E., Wolfe, R. S. & Woese, C. R. (1977) Classification of methanogenic bacteria by 16S ribosomal RNA characterization. Proc. Natl. Acad. Sci. USA 74, 4537–4541.
Gray, M. W. & Doolittle, W. F. (1982) Has the endosymbiont hypothesis been proven? Microbiol. Rev. 46, 1–42.
Hilario, E. & Gogarten, J. P. (1993) Horizontal transfer of ATPase genes—the tree of life becomes a net of life. BioSystems 31, 111–119.
Iwabe, N., Kuma, K., Hasegawa, M., Osawa, S. & Miyata, T. (1989) Evolutionary relationship of archaebacteria, eubacteria, and eukaryotes inferred from phylogenetic trees of duplicated genes. Proc. Natl. Acad. Sci. USA 86, 9355–9359.
Knoll, A. H. (1994) Proc. Natl. Acad. Sci. USA 91, 6743–6750.
Lam, W. C., Cohen, A., Tsouluhas, D. & Doolittle, W. F. (1990) Genes for tryptophan biosynthesis in the archaebacterium Haloferax (Halobacterium) volcanii. Proc. Natl. Acad. Sci. USA 87, 6614–6618.
Maizels, N. (1994) Proc. Natl. Acad. Sci. USA 91, 6729–6734.
Margulis, L. & Guerrero, R. (1991) Kingdoms in turmoil. New Sci. 129, 46–50.
Margulis, L. (1970) Origin of Eukaryotic Cells (Yale Univ. Press, New Haven, CT).
Mayr, E. (1990) A natural system of organisms. Nature (London) 348 , 491.
Meile, L., Stettler, R., Banholzer, R., Kotik, M. & Leisinger, T. (1991) Tryptophan gene cluster of Methanobacterium thermoautotrophicum Marburg: Molecular cloning and nucleotide sequence of a putative trpEGCBAD operon. J. Bacteriol. 173, 5017–5023.
Olsen, G. J., Woese, C. R. & Overbeek, R. (1994) The winds of (evolutionary) change: Breathing new life into microbiology. J. Bacteriol. 176, 1–6.
Ramirez, C., Kopke, A. K. E., Yang, C.-C., Boeckh, T. & Matheson, A. T. (1993) Chapter 14: The structure, function and evolution of archaeal ribosomes. In The Biochemistry of Archaea (Archaebacteria) , eds. Kales, M., Kushner, D. J. & Matheson, A. T. (Elsevier, Amsterdam), pp. 439–466.
Schopf, J. W. (1994) Proc. Natl. Acad. Sci. USA 91, 6735–6742.
Schwartz, R. M. & Dayhoff, M. O. (1978) Origin of prokaryotes, eukaryotes, mitochondria and chloroplasts. Science 199, 395–403.
Simpson, G. G. (1944) Tempo and Mode in Evolution (Columbia Univ. Press, New York), pp. xxix-xxx.
Smith, M. W., Feng, D.-F. & Doolittle, R. F. (1992) Evolution by acquisition: the case for horizontal gene transfer. Trends Biochem. Sci. 17, 489–493.
Sogin, M. C. (1991) Early evolution and the origin of eukaryotes. Curr. Opin. Genet. Dev. 1, 457–463.
Stanier, R. Y. & van Niel, C. B. (1941) The main outlines of bacterial classification. J. Bacteriol. 42, 437–466.
Stanier, R. Y. & van Niel, C. B. (1962) The concept of a bacterium. Arch. Microbiol. 42, 17–35.
Stanier, R. Y. (1970) Some aspects of the biology of cells and their possible evolutionary significance. Symp. Soc. Gen. Microbiol. 20, 1–38.
Stein, D. B. & Searcy, K. B. (1981) A microplasma-like archaebacterium possibly related to the nucleus and cytoplasm of eukaryotic cells. Ann. N.Y. Acad. Sci. 361, 312–323.
Tutino, M. L., Scarano, G., Marino, G., Sannia, G. & Cubellis, M. V. (1993) Tryptophan biosynthesis genes trpEGC in the thermoacidophilic archaebacterium Sulfolobus solfataricus. J. Bacteriol. 175, 299–302.
Woese, C. R. & Fox, G. E. (1977) Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc. Natl. Acad. Sci. USA 74, 5088–5090.
Woese, C. R. (1982) Archaebacteria and cellular origins: an overview. Zbl. Bakt. Hyg. I. Abt. Urig. C3, 1–17.
Woese, C. R. (1987) Bacterial evolution. Microbiol. Rev. 51, 221–271.
Woese, C. R., Kandler, O. & Wheelis, M. L. (1990) Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 87, 4576–4579.
Zillig, W., Palm, P., Klenk, H.-P., Langer, D., Hüdepohl, U., Hain, J., Lanzendörfer, M. & Holz, I. (1993) Chapter 12: Transcription in archaea. In The Biochemistry of Archaea (Archaebacteria), eds. Kales, M., Kushner, D. J. & Matheson, A. T. (Elsevier, Amsterdam), pp. 367–391.
Zillig, W., Stetter, K. O., Schnabel, R., Madon, J. & Gierl, A. (1982) Transcription in archaebacteria. Zbl. Bakt. Hyg. I. Abt. Orig. C3, 218–227.